Exosomal ceRNA Microarray Service
- Exosome Isolation: Customized methods are used to isolate exosomes from various biological fluids (serum, plasma, urine, cerebrospinal fluid, etc.) using techniques such as ultracentrifugation, column chromatography, and membrane affinity.
- Quality Control: Exosome purity and integrity are verified using TEM (Transmission Electron Microscopy), NTA (Nanoparticle Tracking Analysis), and Western Blot for exosome-specific markers (CD9, CD63, CD81).
- RNA Extraction: Exosomal RNA, including LncRNA, circRNA, miRNA, and mRNA, is extracted using specialized kits.
- RNA Quality Control: RNA quantity and quality are assessed using Agilent Bioanalyzer to ensure that the extracted RNA meets the required standards for microarray analysis.
- The extracted RNA is fluorescently labeled and hybridized to a microarray chip with specific probes for different RNA types (LncRNA, circRNA, miRNA, mRNA).
- Stringent washing protocols are followed to minimize background noise and ensure high-quality hybridization.
- The microarray is scanned using high-resolution scanning technology to capture fluorescent signals that correspond to RNA expression levels.
- Differential Expression Analysis: Identifies differentially expressed LncRNAs, circRNAs, miRNAs, and mRNAs.
- ceRNA Interaction Analysis: Predicts miRNA binding sites and explores the regulatory relationships between competing RNAs.
- Functional Enrichment: GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis to explore the functional implications of the identified ceRNA network.
- Raw Data: Full datasets, including differential expression data and ceRNA interaction scores.
- Visualizations: Heatmaps, volcano plots, pathway diagrams, and network maps.
- Final Report: A comprehensive data analysis report providing conclusions, visualizations, and actionable insights.
Exosomes, extracellular vesicles that are present in various bodily fluids, have emerged as key players in cell-to-cell communication and disease progression. These small vesicles carry a variety of biomolecules, including RNA, proteins, lipids, and other active components. Among the RNA species found in exosomes are long non-coding RNAs (LncRNA), circular RNAs (circRNA), microRNAs (miRNA), and messenger RNAs (mRNA). These RNA molecules play crucial roles in gene regulation, cellular processes, and disease mechanisms.
The ceRNA (competing endogenous RNA) hypothesis suggests that LncRNAs, circRNAs, mRNAs, and other non-coding RNAs regulate each other by competing for binding to shared miRNAs, thus influencing gene expression. This competitive relationship forms a complex regulatory network, and understanding these interactions in exosomes is essential for exploring disease mechanisms, identifying molecular biomarkers, and developing novel therapeutic approaches.
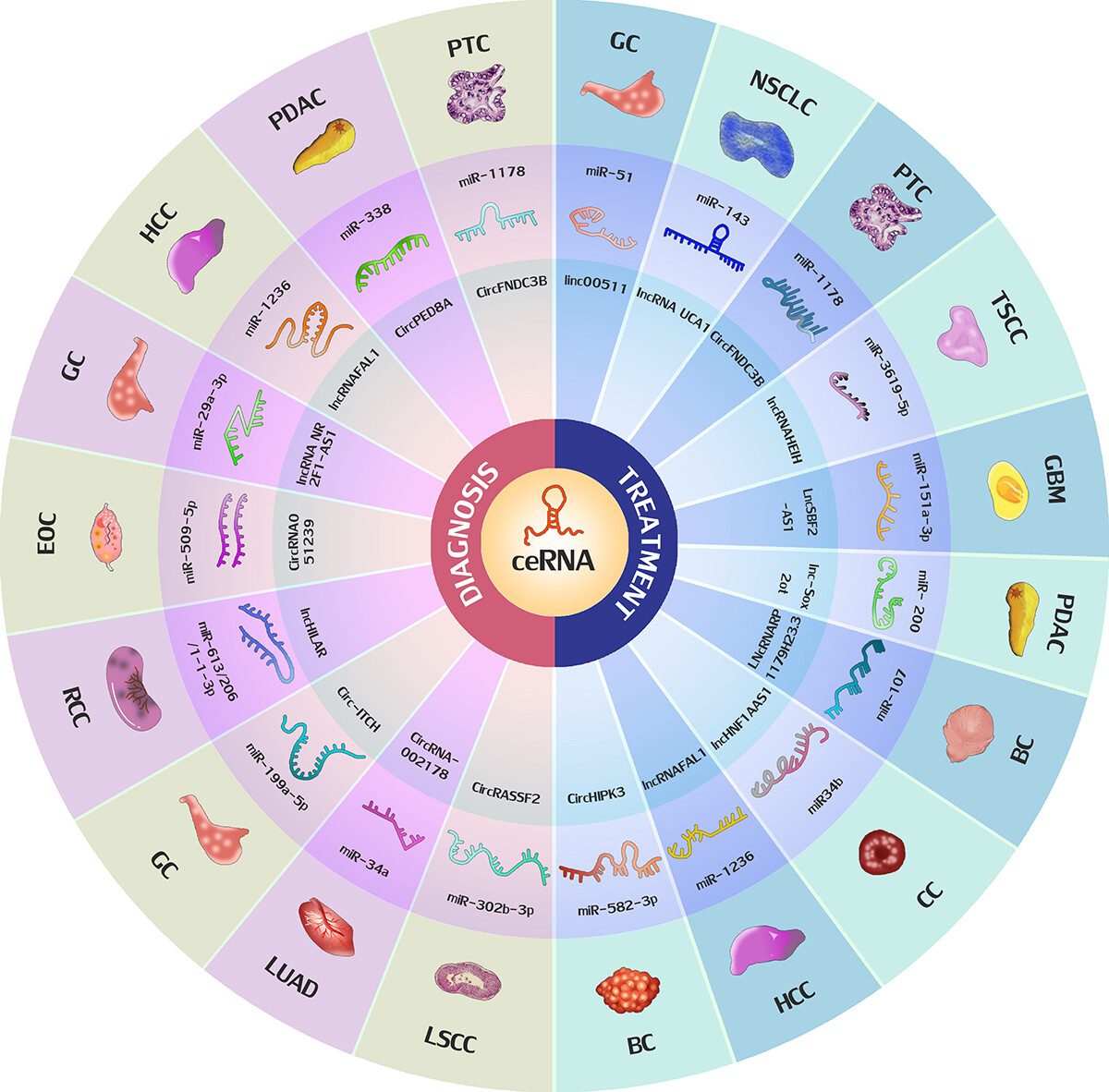
Mao, M. et al. Bioengineered. 2022.
The Role of Exosomal ceRNA in the Diagnosis and Treatment of Malignant Tumors
Service at MtoZ Biolabs
Leveraging a globally leading exosome genomics platform and advanced technologies such as high-throughput microarray, exosome isolation and purification, fluorescent labeling, differential analysis, and ceRNA network modeling, MtoZ Biolabs' Exosomal ceRNA Microarray Service offers a comprehensive, end-to-end solution that provides a full range of services from exosome isolation and ceRNA detection to in-depth network analysis, systematically deciphering the competitive interaction networks of LncRNA, circRNA, miRNA, and mRNA in exosomes, helping researchers accurately identify disease biomarkers, molecular targets, and underlying mechanisms within the ceRNA network, driving the advancement of precision medicine and non-invasive diagnostics.
Analysis Workflow
At MtoZ Biolabs, we follow a strict, standardized workflow to ensure high-quality data output at each step of the analysis:
1. Exosome Isolation & Quality Control
2. RNA Extraction & Quality Check
3. Fluorescent Labeling & Microarray Hybridization
4. Microarray Scanning & Data Collection
5. Bioinformatics Analysis & ceRNA Network Construction
6. Data Delivery & Reporting
Service Advantages
✅ Comprehensive Exosomal ceRNA Profiling: Analyze LncRNAs, circRNAs, miRNAs, and mRNAs together in a single, high-throughput microarray, offering a complete picture of the exosomal ceRNA network.
✅ Optimized for Complex Samples: Tailored exosome isolation and RNA extraction methods ensure high-quality data from a wide range of biological fluids, including low-concentration samples.
✅ Advanced Bioinformatics Support: In-depth bioinformatics analysis including differential expression, ceRNA interaction modeling, and functional enrichment helps to interpret complex regulatory networks.
✅ Reliable Data for Scientific Publication: We provide publication-ready data, with visualizations and detailed reports that support high-quality scientific publications and grant applications.
✅ Customizable Solutions: We offer flexible solutions for different research needs, including integration with other omics analyses (proteomics, metabolomics) for comprehensive systems biology studies.
Applications
1. Cancer Biomarker Discovery
Uncover exosomal ceRNA networks in cancers to identify non-invasive molecular biomarkers for early detection, diagnosis, and prognosis.
2. Mechanistic Studies in Neurodegenerative Diseases
Explore the roles of exosomal ceRNA in diseases like Alzheimer’s, Parkinson’s, and Huntington’s to understand molecular mechanisms and potential therapeutic targets.
3. Cardiovascular Disease Research
Analyze exosomal ceRNA in heart diseases, such as myocardial infarction and atherosclerosis, to identify biomarkers and regulatory pathways.
4. Immunological and Inflammatory Disease Studies
Study the role of exosomal ceRNA in autoimmune diseases and inflammation, uncovering new regulatory mechanisms for immune modulation.
5. Drug Mechanism and Resistance Research
Investigate the role of ceRNA networks in drug resistance and the pharmacodynamics of therapeutic agents to guide personalized treatment strategies.
6. Cell-to-Cell Communication & Microenvironment Regulation
Examine exosomal ceRNA in cell communication within the tumor microenvironment or stem cell niches, uncovering new therapeutic insights.
FAQ
Q1: What types of samples are suitable for Exosomal ceRNA microarray analysis?
A: MtoZ Biolabs’ Exosomal ceRNA microarray service is suitable for a wide variety of biological fluids, including serum, plasma, urine, saliva, cerebrospinal fluid, and amniotic fluid. Our protocols are customized to ensure optimal exosome isolation and RNA extraction from different sample types.
Q2: How should exosomal samples be stored and transported?
A: It is recommended to use RNase-free cryogenic vials, freeze rapidly in liquid nitrogen, and store at -80°C. During transportation, dry ice should be used to maintain temperature, and repeated freeze-thaw cycles should be avoided.
Q3: Can dynamic changes of ceRNA across different disease stages be analyzed?
A: Yes, we support multi-group comparative analysis (e.g., healthy controls vs. early-stage patients vs. late-stage patients).
How to order?







