Epigenomics (Chromatin Microenvironment) Analysis Service
- Cancer Research: Investigate the roles of epigenetic modifications in tumor initiation, progression, and drug resistance, identifying potential therapeutic targets.
- Immunology: Explore how epigenetic regulation affects immune cell development and function, providing insights into immune disorders and autoimmune diseases.
- Neuroscience: Study epigenetic regulation in neural development and neurodegenerative diseases, uncovering key mechanisms in neural dysfunction.
- Environmental Epigenetics: Assess how environmental factors influence epigenetic modifications, providing insights into environmental diseases and susceptibility.
Epigenomics focuses on studying the regulatory mechanisms that influence gene expression without altering the underlying DNA sequence. These mechanisms, which include DNA methylation, histone modifications, chromatin remodeling, and non-coding RNA regulation, play critical roles in controlling gene expression, driving cell differentiation, guiding development, and contributing to disease pathology. The chromatin microenvironment significantly impacts gene accessibility, and thus, epigenomic analysis is essential for uncovering the molecular mechanisms that govern gene regulation, particularly in the context of diseases such as cancer, neurodegenerative disorders, and immune dysfunctions.
By examining chromatin states, DNA modifications, and histone markers, researchers can gain a detailed understanding of the genome's dynamic behavior and how it responds to environmental cues and biological signals. MtoZ Biolabs offers comprehensive Epigenomics Analysis Service that provides deep insights into chromatin microenvironment regulation, aiding researchers in advancing studies on disease mechanisms and therapeutic approaches.
Services at MtoZ Biolabs
MtoZ Biolabs' epigenomics analysis platform leverages state-of-the-art technologies, including chromatin immunoprecipitation sequencing (ChIP-seq), ATAC-seq, and bisulfite sequencing for DNA methylation studies. We offer full-scale solutions that encompass DNA methylation profiling, histone modification analysis, and chromatin accessibility evaluation. Using these advanced techniques, we enable clients to comprehensively study alterations in the chromatin microenvironment, uncovering the epigenetic mechanisms underlying gene expression regulation.
Analysis Workflow
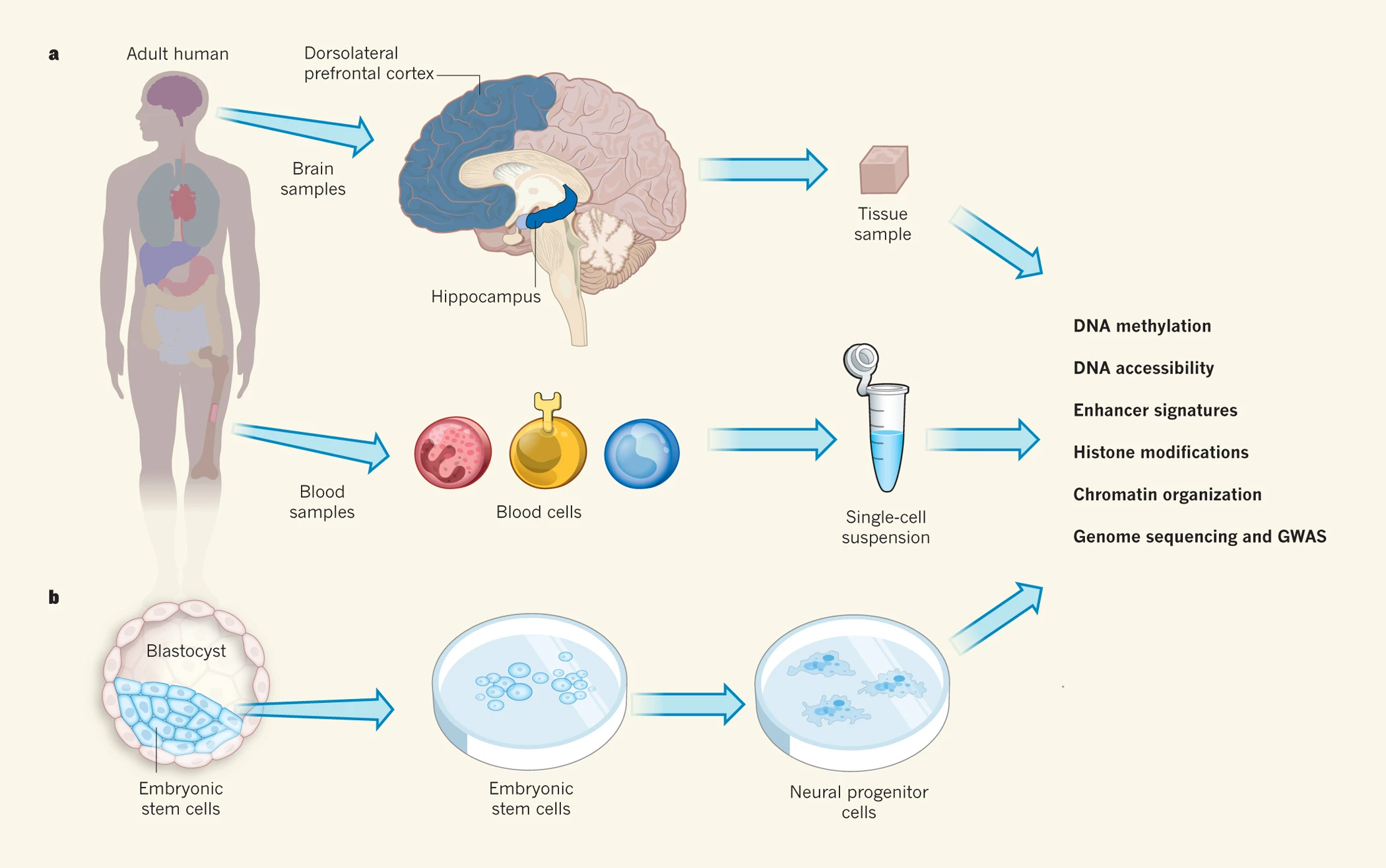
Romanoski, C. E. et al. Nature. 2015.
Service Advantages
Why Choose MtoZ Biolabs?
1. Extensive Epigenetic Profiling: We provide thorough epigenomics services, covering chromatin accessibility, histone modifications, and DNA methylation to reveal intricate gene regulatory networks.
2. High Sensitivity and Precision: Our use of cutting-edge sequencing technology ensures the accurate detection of even low-abundance epigenetic markers, delivering highly reproducible results.
3. Customizable Solutions: Our services are tailored to meet the specific research objectives of each client, offering personalized experimental designs and data analysis plans.
4. Rapid Turnaround Time: Our streamlined workflows and efficient data processing allow us to deliver high-quality results promptly, supporting the timely progression of research.
Sample Submission Requirements
1. Sample Types
We accept various sample types, including cells, tissues, or biological fluids (e.g., serum).
2. Sample Volume
For cell samples, 5-10 million cells are recommended, while tissue samples should be at least 100-200 mg.
3. Sample Preservation
Store samples at -20°C or lower to prevent degradation, avoiding repeated freeze-thaw cycles.
Applications
Deliverables
What Could be Included in the Report?
1. Comprehensive Experimental details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
How to order?







