Edman Sequencing vs. Mass Spectrometry: A Comparison of Protein Sequencing Techniques
Edman Sequencing and Mass Spectrometry (MS) are two of the most widely employed protein sequencing techniques for elucidating the primary structure of proteins. Edman sequencing involves stepwise chemical cleavage, whereas Mass Spectrometry utilizes ionization and fragmentation processes followed by analysis of the resulting mass-to-charge ratios. This paper presents a detailed comparison of these two methods, aiming to clarify their respective strengths, limitations, and appropriate applications.
Comparison of Technical Principles
1. Edman Sequencing: Sequential Cleavage via Chemical Reactions
(1) Core Mechanism: The process involves coupling phenyl isothiocyanate (PITC) to the α-amino group at the N-terminus of a polypeptide, followed by intramolecular cyclization, cleavage, and iterative identification of the liberated amino acid residues.
(2) Information Acquisition: Enables direct determination of the N-terminal sequence without reliance on existing databases or prior knowledge, thereby constituting a form of de novo sequencing.
(3) Modification Analysis: Capable of detecting modifications specific to the N-terminus (e.g., acetylation), but unable to identify internal modifications such as phosphorylation.
2. Mass Spectrometry: Comprehensive Analysis through Ionization and Fragmentation
(1) Core Mechanism: Peptide fragments are ionized and analyzed using a mass analyzer to determine their mass-to-charge ratios (m/z). Subsequent fragmentation via techniques like collision-induced dissociation (CID) generates tandem mass spectra, from which the peptide sequence can be inferred.
(2) Information Acquisition: Typically relies on matching spectra to database entries (e.g., UniProt); for novel sequences or uncharacterized organisms, de novo sequencing algorithms are necessary.
(3) Modification Analysis: Allows global detection of various types of modifications—including those at the N-terminus, C-terminus, and internal residues—though prior specification of expected modifications or the application of open search strategies is often required to broaden the scope of detectable changes.
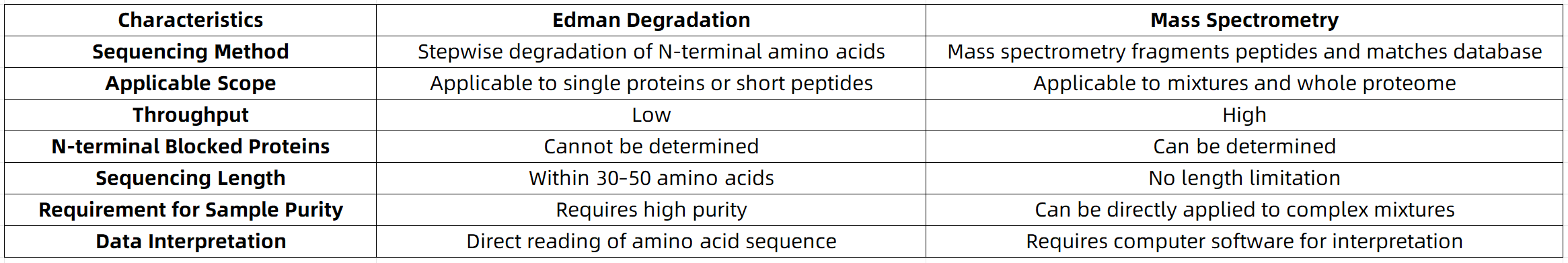
Intuitive Comparison of Performance Parameters Between Edman Sequencing and Mass Spectrometry
Comparison of Application Scenarios for Protein Sequencing Techniques
1. Advantageous Scenarios of Edman Sequencing
(1) N-terminal sequence verification: Assessment of N-terminal sequence consistency in recombinant proteins for regulatory compliance in biopharmaceutical applications;
(2) High-precision sequencing of short peptides: Applicable to the analysis of antigenic epitopes, neuropeptides, or proteolytic fragments;
(3) De novo sequencing of unknown samples: Suitable for ancient proteins or proteins from newly identified species in the absence of reference databases;
(4) Direct identification of N-terminal modifications: Enables confirmation of modifications such as N-terminal acetylation and pyroglutamylation.
2. Advantageous Scenarios of Mass Spectrometry
(1) High-throughput omics applications: Large-scale identification and quantitative analysis of proteins;
(2) Comprehensive sequence coverage: Reconstruction of full-length protein sequences through overlapping peptide mapping;
(3) Profiling of post-translational modifications: Global characterization of modifications including phosphorylation and glycosylation;
(4) Analysis of complex biological samples: Effective for identifying proteins in mixtures such as serum or tissue lysates.
3. Cross-Application Scenarios
(1) Elucidation of challenging samples: Edman degradation can be employed to confirm unknown peptide sequences identified by mass spectrometry;
(2) Precision diagnostics: Edman degradation verifies disease-associated short peptide biomarkers, while mass spectrometry facilitates high-throughput screening;
(3) Paleoproteomics research: Edman degradation enables characterization of severely degraded short peptides, with mass spectrometry providing complementary internal sequence information.
Edman sequencing and mass spectrometry each offer distinct advantages and are suited to different scenarios in protein sequencing. Edman degradation is ideal for precise N-terminal sequencing of highly purified individual proteins, whereas mass spectrometry has emerged as a central technique in modern proteomics due to its high throughput, rapid detection, and broad applicability. Depending on experimental requirements, researchers may select the most appropriate method or integrate both approaches to achieve more comprehensive and accurate protein sequence analysis. MtoZ Biolabs is dedicated to delivering advanced, high-quality protein N-terminal sequencing services based on Edman degradation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







