Edman Degradation Sequencing Services
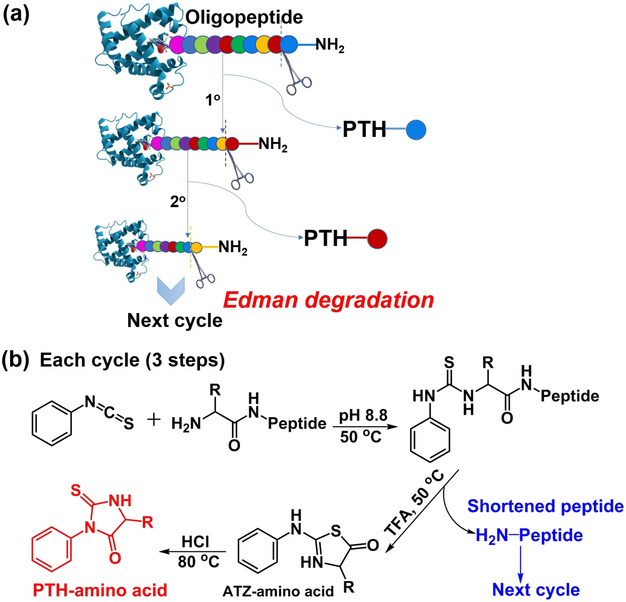
The Edman degradation sequencing services use the Edman degradation method to determine the N-terminal amino acid sequence in the peptide chain or protein, that is, through the reaction of PITC (phenylene isothiocyanate) and the α-amino group at the N terminus of the protein or polypeptide. The first amino acid at the N-terminus is broken off from the sequence to obtain the free PTH-amino acid (Figure 1). The generated PTH amino acid is analyzed by high performance liquid chromatography, and the remaining polypeptide protein sequence undergoes the above reaction in sequence to generate Corresponding PTH-amino acid. The peptide/protein sequence is determined by repeating these processes until the peptide/protein is completely degraded. For proteins (polypeptides) with longer sequences, you can also cut the sample into small peptide segments first, analyze the peptide sequence first, and then splice the peptide information to obtain the sequence information of the complete protein.
With the advancement of the modern pharmaceutical industry, a large number of protein- and peptide-based drug molecules have emerged. The N-terminal amino acid sequence of proteins is a critical quality attribute of biopharmaceuticals. Furthermore, the biosynthesis of proteins begins at the N-terminus, and the composition of the N-terminal sequence has a profound impact on biological functions, such as protein half-life and subcellular localization. In addition, various post-translational modifications (PTMs) can occur at the N-terminus, which are closely associated with protein function and stability.As a routine method of analyzing n-terminal sequence of protein, Edman degradation sequencing service can help researchers to analyze the high-level structure of protein and reveal the biological function of protein.
Zhang, H. et al. Chemistry. 2020.
Figure 1. (a) Chemical Reaction of N-terminal Sequencing Based on Edman Degradation (b) Detailed Reaction Steps
Services at MtoZ Biolabs
The Edman degradation sequencing services provided by MtoZ Biolabs uses the PPSQ series instruments launched by Shimadzu Corporation to analyze the N-terminal amino acid sequence of protein or peptide samples, and can analyze more than 30 N-terminal amino acids.
Service Advantages
1. Advanced Analytical Platform
MtoZ Biolabs has established an advanced Edman degradation sequencing services platform to ensure reliable, fast and high-precision analysis services.
2. Transparent Pricing
Our pricing is transparent with no hidden fees or extras.
3. High Data Quality
Data quality control is strict and comprehensive data reports are provided to customers.
4. Customized Research Solutions
MtoZ Biolabs offers tailored services based on sample type to address unique research questions and experimental requirements.
Sample Requirements
1. Sample Purity
The purity of the sample must be >90%, the salt ion content should be within 50 mmol/L, it should not contain denaturants such as SDS, and the N-terminal must be a uniform, high-purity sample.
2. Sample Quantity
10 pmol sample can measure 20 aa.
3. Liquid Samples
Liquid samples cannot contain proteases, and it is recommended to test as early as possible to prevent them from being easily degraded if it takes too long.
4. Membrane-Transferred Samples
The transmembrane samples should not be left for too long before transfer to prevent sample diffusion. After transfer, the samples can be sealed and stored for 3-6 months. It is recommended that samples should be stained with Ponceau or Coomassie R-250, and CAPS buffer should be used for electrophoresis buffer.
*Note: If you have special requirements or need assistance with sample preparation, please contact us.
Applications
1. Biopharmaceutical research and development
In biopharmaceutical development, Edman degradation sequencing services are used to determine the sequence of protein-based drugs, verifying their integrity and purity to ensure compliance with quality standards. Additionally, this method can be applied to study the degradation pathways and metabolic routes of protein drugs, providing crucial insights into their stability and pharmacokinetics.
2. Protein Structure and Function Studies
By using Edman degradation sequencing services to determine protein sequences, researchers can uncover the relationship between protein structure and function, helping scientists understand the specific roles and interaction mechanisms of proteins within organisms.
3. Protein Engineering and Mutation Analysis
By using Edman degradation sequencing service to determine the sequence of mutated proteins, evaluate the impact of mutations on protein structure and function, and further guide the optimization and improvement of protein engineering.
Case Study
Edman degradation sequencing services have assisted researchers in uncovering a novel immune response mechanism against Group A Streptococcus (GAS) infections: the GAS virulence factor SpeB activates GSDMA through cleavage, triggering pyroptosis in skin epithelial cells and inhibiting systemic infection.
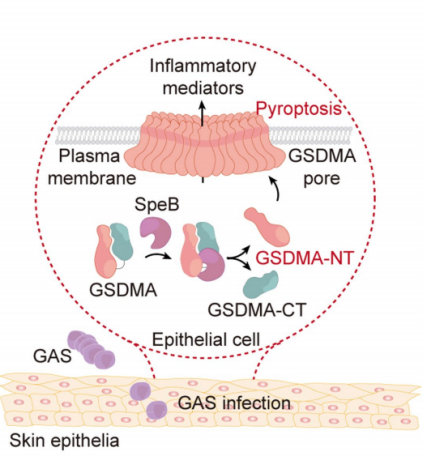
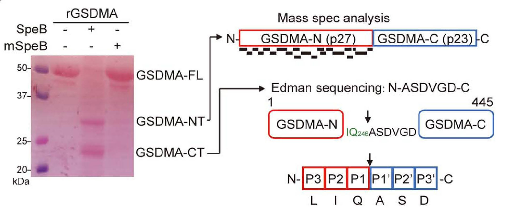
Group A Streptococcus (GAS), as a common pathogenic bacterium, can causes various acute inflammatory diseases. One of its critical virulence factors, streptococcal pyrogenic exotoxin B (SpeB), facilitates GAS colonization on epithelial surfaces and systemic spread. However, the exact mechanism of SpeB's function remained unclear.Researchers aimed to identify how SpeB induces pyroptosis, a form of programmed cell death. Using a CRISPR/Cas9 screening system, they explored potential SpeB-dependent host genes in A431 cells. Among the identified candidates, only the knockout of GSDMA replicated the screening results. The researchers discovered that SpeB can specifically cleave GSDMA (but not other members of the Gasdermins family). The author then used the Edman degradation sequencing services to find the SpeB cleavage site located after Q246. This cleavage behavior occurred in the linker region of GSDMA, causing GSDMA to release two truncation bodies, the N-terminal and the C-terminal, of which only GSDMA-N can directly Causes cell pyroptosis. This case demonstrates how Edman degradation sequencing services can help scientific researchers reveal novel immune response mechanisms against Group A Streptococcus (GAS) infections, elucidating disease mechanisms, and guiding the development of therapeutic strategies.
Deng, W. et al. Nature. 2022.
Figure 2. Model of GAS Virulence Factor SpeB Activating GSDMA to Induce Pyroptosis in Skin Epithelial Cells
Deng, W. et al. Nature. 2022.
Figure 3. Analysis of GSDMA N-terminal and C-terminal Cleavage Products (p27 and p23) by Mass Spectrometry (MS) and Edman Sequencing
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Chromatograms & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







