DIA+PRM Target Proteomics Services
DIA+PRM Target Proteomics Services combine two advanced mass spectrometry technologies, Data-Independent Acquisition (DIA) and Parallel Reaction Monitoring (PRM), to provide high-sensitivity, high-specificity targeted protein analysis. DIA is a full-scan mode that can detect all ion signals simultaneously and resolve peptide information through high-resolution mass spectrometry data, making it particularly suitable for proteomic analysis in complex samples. PRM is a targeted quantitative technique that enhances quantification precision and specificity by selectively detecting fragment ions of specific peptides. It is ideal for accurate quantification of low-abundance proteins and can serve as an alternative to Western blotting and ELISA.
DIA+PRM Target Proteomics Services leverage DIA to provide global proteomic data, allowing the identification of potential target proteins, while PRM further focuses on precise verification and quantification of target proteins, enabling a seamless transition from global screening to accurate quantification. Supported by advanced mass spectrometry platforms and extensive technical expertise, MtoZ Biolabs' DIA+PRM Target Proteomics Services achieve a perfect combination of high data coverage and high-precision quantification, providing solid data support for scientific research.
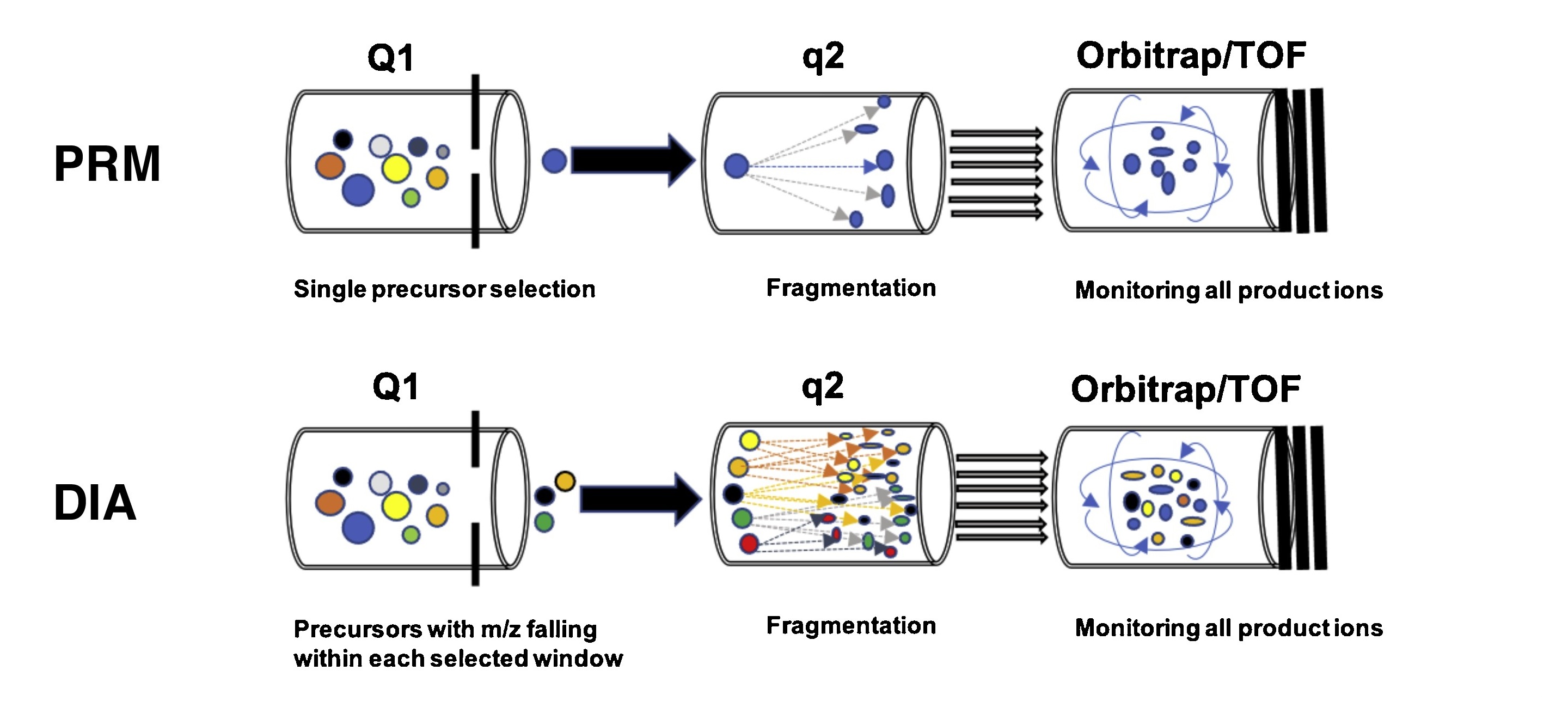
Li J. et al. Drug Discovery Today Technologies. 2021.
Analysis Workflow
General workflow of DIA+PRM Target Proteomics Services:
1. Sample Preparation
Standardized protein extraction and digestion procedures are applied to ensure protein integrity and efficient enzymatic digestion. If necessary, specific post-translational modifications (PTMs) are enriched.
2. DIA Data Acquisition
Digested peptides are separated using high-performance liquid chromatography (HPLC). Mass spectrometry is performed in DIA mode, capturing full-spectrum data across pre-defined windows to generate comprehensive proteome-wide quantitative data.
3. DIA Data Analysis
Specialized software is used for data processing, quantitative analysis, and candidate target screening. Database searches and bioinformatics analysis are conducted to identify protein variations related to the research objectives.
4. PRM Targeted Validation
Set predetermined precursor and product ion transitions for candidate proteins, and perform high-resolution targeted data acquisition in PRM mode. Use software to analyze data, obtain accurate quantitative results, and confirm the expression level and dynamic changes of candidate proteins.
5. Data Integration and Biological Interpretation
Integrate DIA and PRM data to perform pathway enrichment, network analysis and functional annotation. Output detailed reports.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced DIA+PRM Target Proteomics Services platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all DIA+PRM Target Proteomics Services data, providing clients with a comprehensive data report.
4. Comprehensiveness and precision: DIA performs global screening and PRM performs targeted verification to ensure accurate quantification of target proteins.
5. Combination of high throughput and high sensitivity: DIA is suitable for large-scale proteome screening, while PRM can accurately quantify low-abundance target proteins.
Applications
Biomarker discovery and validation
For example, in disease diagnosis, prognosis assessment, and treatment response monitoring, DIA+PRM Target Proteomics Services can be used to discover and validate low-abundance or key regulatory proteins.
Disease mechanism research
Reveal the dynamic changes of protein expression and modification in complex biological samples, and analyze signaling pathways and regulatory networks.
Drug target analysis
Analyze the drug action mechanism, identify key proteins regulated by drugs, and provide quantitative data support for new drug development.
FAQ
Q. Why choose to combine DIA and PRM?
DIA provides high throughput and global proteome coverage, which is suitable for screening potential target proteins from complex samples, and can capture low-abundance proteins and obtain extensive quantitative information. PRM can perform high-sensitivity verification and precise quantification of candidate proteins screened by DIA, improve quantitative specificity and accuracy, and is especially suitable for accurate analysis of low-abundance proteins. Combining DIA and PRM can complement each other's advantages and disadvantages and maximize data accuracy and coverage.
Q. In what situations is the combination of DIA and PRM the best?
During the screening and accurate quantification of target protein groups, especially when the research requires high-coverage whole-proteome quantification and further verification of certain candidate proteins.
When analyzing complex samples, such as clinical samples or small sample volumes, combining the high throughput of DIA with the high sensitivity of PRM can improve the reliability of research results while ensuring breadth and accuracy.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized a high-throughput mass spectrometry strategy combining DIA and PRM to comprehensively screen the serum proteome of hepatocellular carcinoma (HCC) patients and healthy controls, followed by targeted validation of candidate biomarkers. Using DIA technology, researchers initially quantified and identified a large number of serum proteins associated with early-stage HCC development. PRM technology was then employed for high-precision quantification of target proteins with potential diagnostic value. The results revealed significant differences in specific proteins between HCC patients and controls, providing reliable biomarker candidates for non-invasive early diagnosis of HCC.
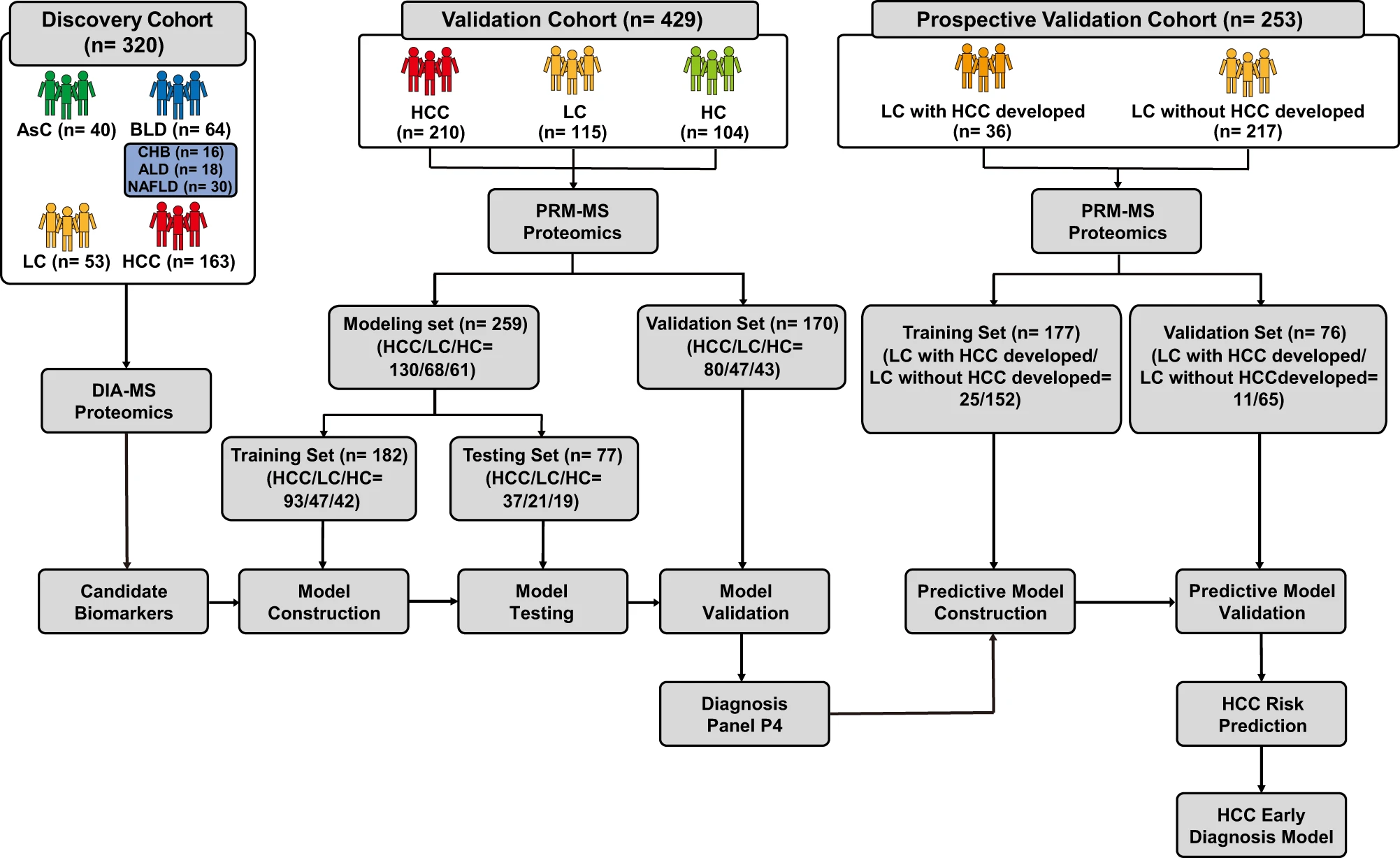
Xing X. et al. Nature Communications. 2023.
How to order?







