De Novo Protein Sequencing: How to Decode Unknown Proteins?
-
Previously unannotated proteins
-
Novel splice variants
-
Pathogen-derived, tumor-specific, or exogenously expressed proteins
-
Sequence deviations introduced by post-translational modifications or mutations
-
Use of multi-enzyme digestion strategies (e.g., Trypsin combined with Chymotrypsin) to generate overlapping peptide fragments
-
Acquisition of high-resolution MS/MS data using instruments such as the Orbitrap Fusion Lumos
-
Manual spectrum validation and structural modeling to correct potential misassemblies
-
Application of multi-round digestion combined with enrichment techniques (e.g., high-pH reverse-phase chromatography for peptide pre-fractionation)
-
Integration of multi-omics data, such as transcriptomic validation
-
Employment of inference-based assembly algorithms and sequence similarity scoring models to enhance sequence reconstruction quality
-
Homology-based analysis using tools such as BLAST to infer structure and functional domains
-
In silico prediction of secondary and tertiary structures to assess potential functional categories, such as enzymes, signaling proteins, or antigens
-
Experimental validation of expression and function, including activity assays and ELISA-based methods
-
High-resolution mass spectrometry using Orbitrap Fusion Lumos and timsTOF Pro
-
Multi-enzyme digestion combined with fractionation-based enrichment
-
Parallel processing through widely adopted De Novo sequencing algorithms such as PEAKS, pNovo, and Novor
-
In-house developed modules for sequence assembly optimization and post-translational modification integration
-
Homology-based annotation and functional scoring systems specifically designed for unknown proteins
-
Manual verification of MS/MS results by a team of PhD-level scientists for every project
-
Full-length De Novo protein sequences
-
Peptide coverage maps with annotated modifications
-
BLAST alignment reports accompanied by functional prediction annotations
-
Optional services including expression validation and bioactivity assays
In proteomics, clinical sample analysis, and studies involving non-model organisms, researchers increasingly face a recurring challenge: peptide fragments identified through mass spectrometry fail to match any known entries in existing databases. These sequences, termed “unknown proteins” or “orphan peptides,” may originate from:
Conventional database-dependent search tools (e.g., Mascot, MaxQuant), which rely on precompiled FASTA reference databases, exhibit limited capability in identifying such proteins. In these cases, De Novo protein sequencing emerges as an essential approach.
What Is De Novo Protein Sequencing? How Does It Differ from Database Search?
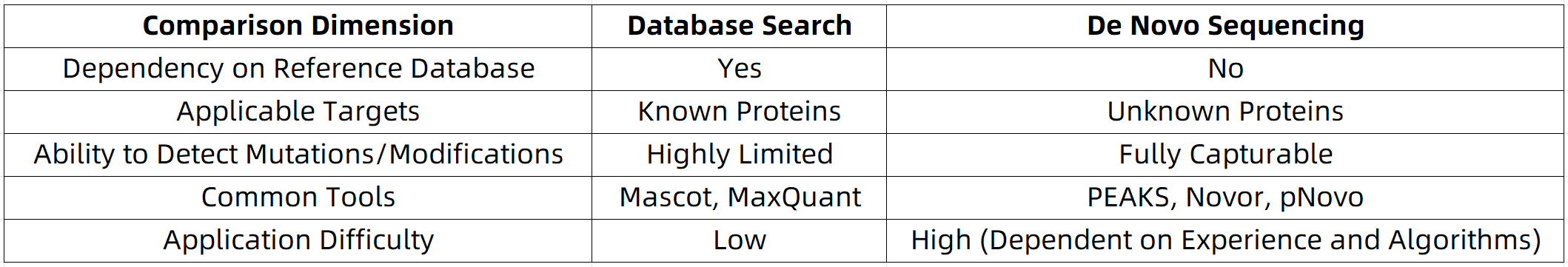
De Novo sequencing (also known as “from scratch” sequencing) refers to a method that directly deduces protein amino acid sequences from MS/MS fragmentation data without relying on any pre-existing sequence database. Compared to database search methods, key differences include:
What Types of Research Require De Novo Sequencing to Decode Unknown Proteins?
1. Research on Non-Model Organisms
In studies involving well-established model organisms such as mice and humans, protein databases are typically comprehensive and well-annotated. However, for non-model organisms—including various fish species, plants, and microorganisms—genomic data are often incomplete or inaccurately annotated, resulting in low identification rates in database-driven searches. Therefore, De Novo sequencing is essential for obtaining accurate protein sequences in such cases.
2. Tumor Neoantigen Screening
Mutated or fusion-derived proteins often contain single amino acid alterations that prevent their identification through standard database searches. De Novo sequencing enables direct detection of these mutation sites at the mass spectrometry level, making it a critical tool in cancer immunotherapy for identifying novel neoantigens.
3. Characterization of Proteins from Exogenous Expression or Unknown Sources
In complex biological products such as traditional medicine formulations, natural extracts, or recombinant expression systems, the exact protein composition is often unknown, and no corresponding genomic information is available. In such scenarios, De Novo sequencing provides the only viable route for structural characterization of these proteins.
4. Identification of Post-Translationally Modified Proteins
Standard database-based search methods struggle to identify peptide sequences that contain novel or uncharacterized post-translational modifications. De Novo sequencing, when combined with specialized modification-detection algorithms, allows simultaneous determination of both peptide sequences and their corresponding modification sites.
What Are the Key Technical Challenges in Decoding Unknown Proteins?
Challenge 1: Complex Fragmentation Patterns Complicate Algorithmic Interpretation
Mass spectrometry data often exhibit complications such as neutral loss, co-elution of isomeric amino acids (e.g., Isoleucine/Leucine), and overlapping signals from post-translational modifications. These factors hinder accurate peptide assembly by automated algorithms.
MtoZ Biolabs Solutions:
Challenge 2: Insufficient Sequence Coverage Prevents Full-Length Reconstruction
Low expression levels or inefficient enzymatic digestion of certain proteins may lead to insufficient peptide coverage, making it difficult to reconstruct the complete protein sequence.
MtoZ Biolabs Solutions:
Challenge 3: Lack of Functional Annotation Impedes Protein Identification
Even with a successfully reconstructed sequence, determining the protein’s function remains a significant challenge in the absence of annotation.
MtoZ Biolabs Solutions:
How Does MtoZ Biolabs Facilitate the Structural Elucidation of Unknown Proteins?
We offer a comprehensive De Novo protein sequencing workflow, from sample preparation to functional validation, including:
1. Technical Platform Support
2. Proprietary Algorithms and Expert Review
3. Deliverables
As increasingly complex biological samples, non-model organisms, and mutated proteins become central to current research, De Novo protein sequencing is evolving from a niche technology into an indispensable tool for advancing life sciences. Leveraging a robust mass spectrometry platform and extensive experience in unknown protein characterization, MtoZ Biolabs provides dependable support for your research—enabling the decoding of uncharacterized proteins from experimental data and revealing novel functions through structural insights. If you are working on the identification of unknown proteins or aim to characterize the structure of unannotated protein therapeutics, we welcome you to contact the MtoZ Biolabs technical team for sample evaluation and customized sequencing strategies.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







