Comprehensive Overview of Mass Spectrometry-Based Protein Phosphorylation Analysis Technologies
Protein phosphorylation is one of the most prevalent and functionally significant post-translational modifications, playing essential roles in a wide range of biological processes, including cell proliferation, differentiation, metabolism, and apoptosis. Precise identification and quantitative characterization of phosphorylation sites enable researchers to systematically decipher the dynamic regulation of cellular signaling networks, elucidate molecular mechanisms underlying disease pathogenesis, and identify potential therapeutic targets. Among the various analytical approaches available, mass spectrometry-based phosphoproteomics has emerged as a central technology in modern proteomics due to its high throughput, sensitivity, and capability for molecular-level resolution.
How Does Mass Spectrometry Detect Protein Phosphorylation?
1. Ionic Characteristics Underlying Phosphorylation Detectability
The incorporation of a phosphate group substantially alters the physicochemical properties of peptides, resulting in a mass increase of 79.966 Da. In positive ion mode, phosphorylated peptides are prone to characteristic neutral loss events (loss of H₃PO₄, 98 Da). These features constitute a fundamental basis for the mass spectrometric detection and identification of phosphorylation.
2. Major Mass Spectrometry Platforms and Fragmentation Strategies
State-of-the-art mass spectrometry platforms, such as Orbitrap Fusion Lumos and timsTOF Pro 2, combined with multiple fragmentation techniques (CID, HCD, and ETD), enable highly sensitive and reliable identification of phosphopeptides. Commonly employed strategies include:
(1) CID (Collision-Induced Dissociation): frequently induces neutral loss of phosphate groups, facilitating initial screening of phosphorylated peptides.
(2) ETD (Electron Transfer Dissociation): preserves labile post-translational modifications, making it particularly suitable for precise localization of multiple phosphorylation sites.
(3) HCD (Higher-Energy Collisional Dissociation): generates high-quality b/y ion spectra, supporting both confident identification and quantitative analysis.
Experimental Workflow: Key Steps from Sample Preparation to Data Generation
1. Sample Preparation and Protein Extraction
Because phosphorylation is typically a low-abundance and labile modification, sample handling must be performed with particular care. The inclusion of protease and phosphatase inhibitors effectively minimizes protein degradation and dephosphorylation. Subsequent steps include cell lysis, protein quantification, reduction and alkylation, and enzymatic digestion.
2. Phosphopeptide Enrichment: A Critical Determinant of Analytical Success
Given the extremely low abundance of phosphopeptides within complex proteomic backgrounds, enrichment represents an indispensable step. Commonly used approaches include:
(1) IMAC (Immobilized Metal Affinity Chromatography)
Exploits the affinity between phosphate groups and metal ions such as Fe³⁺ and Ga³⁺.
(2) TiO₂ enrichment (Titanium Dioxide)
Particularly effective for mono-phosphorylated peptides, offering high selectivity.
(3) Antibody-based enrichment (phospho-tyrosine antibodies)
Specifically targets tyrosine-phosphorylated peptides and is widely applied in signaling pathway analyses.
3. LC-MS/MS Analysis and Data Acquisition
Following enrichment, phosphopeptides are introduced into high-resolution mass spectrometers via nano-scale liquid chromatography. Two principal data acquisition strategies are commonly applied: Data-Dependent Acquisition (DDA), which is well suited for the discovery of novel phosphorylation events, and Data-Independent Acquisition (DIA), which enables high-throughput and reproducible quantification of predefined phosphorylation sites.
4. Data Processing and Phosphorylation Site Assignment
Phosphorylation site identification is achieved through database searching using software platforms such as MaxQuant, Proteome Discoverer, and Spectronaut, in combination with site localization scoring algorithms (e.g., Ascore and ptmRS). Downstream analyses, including pathway enrichment and kinase–substrate relationship prediction, facilitate the reconstruction of signaling network architectures.
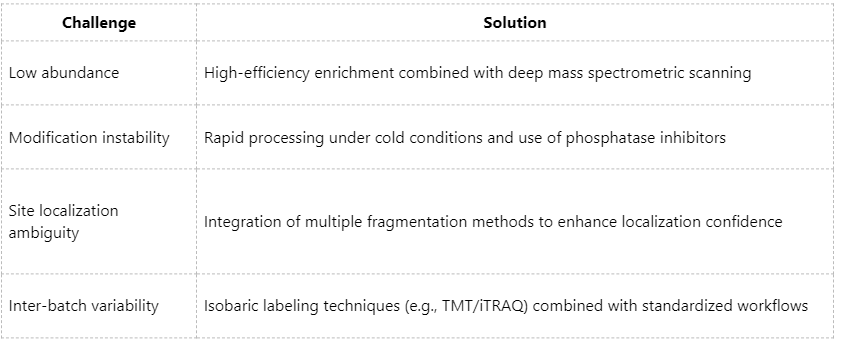
Technical Challenges and Corresponding Solutions
Application Scenarios: From Basic Research to Clinical Translation
1. Investigation of Oncogenic Signaling Pathways
For example, activation profiling of pathways such as PI3K/AKT and MAPK.
2. Drug Target Validation and Kinase Inhibitor Screening
Assessment of pharmacological intervention effects through quantitative phosphorylation analysis.
3. Decoding Immune Responses and Inflammatory Signaling
Dynamic monitoring of coordinated kinase activities across signaling cascades.
4. Biomarker Discovery in Personalized Medicine
Construction of phosphoproteomic landscapes using clinical specimens.
Mass spectrometry-based phosphoproteomic technologies have become a cornerstone of contemporary proteomics research. Through continuous optimization of sample preparation protocols, enrichment strategies, and mass spectrometric acquisition methods, researchers can reliably detect low-abundance and dynamically regulated phosphorylation events within complex biological systems. These advances provide robust analytical foundations for mechanistic studies of disease and the development of targeted therapeutics. If you are engaged in research on cellular signaling pathways, kinases, or the identification of novel drug targets, we invite you to contact MtoZ Biolabs. Our expertise and technical capabilities are designed to support and advance your research objectives.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







