Complexome Profiling Service
Complexome profiling is a high-throughput technique used to systematically study the composition, assembly state, and dynamic changes of protein complexes. This method combines complex separation under mild conditions (such as electrophoresis, size-exclusion chromatography, or density gradient centrifugation) with mass spectrometry analysis to enable precise identification and quantification of complex components, thereby generating distribution and dynamic change profiles under different conditions.
The complexome profiling service is widely applied in the study of multiprotein systems such as the mitochondrial respiratory chain, ribosomes, and membrane protein complexes. It is suitable for investigating complex assembly mechanisms, disease-related conformational alterations, protein interaction network remodeling, and functional regulatory mechanisms. This service provides strong data support for deciphering multiprotein cooperation and disease-related mechanisms.
Services at MtoZ Biolabs
Leveraging high-resolution mass spectrometry platforms, MtoZ Biolabs has launched the complexome profiling service focusing on the analysis of protein complex composition, assembly state, and dynamic characteristics. This service separates native complexes under mild conditions using methods such as electrophoresis, size-exclusion chromatography, or density gradient centrifugation, combined with fractionated mass spectrometry detection to systematically analyze the composition, abundance, and migration features of multiprotein complexes under different conditions. The service generates high-throughput, visualized complex migration profiles and quantitative protein data, supporting studies on protein assembly mechanisms, interaction networks, and disease-related functional abnormalities.
Analysis Workflow
1. Sample Lysis and Complex Extraction
Cells or tissues are lysed under mild conditions to extract protein complexes while preserving their native conformation, avoiding dissociation or denaturation.
2. Complex Separation
Techniques such as electrophoresis, size-exclusion chromatography, or density gradient centrifugation are used to fractionate protein complexes based on size or migration rate.
3. Mass Spectrometry Analysis
Each collected fraction is enzymatically digested and analyzed using high-resolution mass spectrometry for protein identification and quantification.
4. Data Processing and Profile Generation
The distribution of each protein across the gradient is calculated to generate complex migration profiles, followed by in-depth analysis of interaction patterns, assembly states, and functional enrichment.
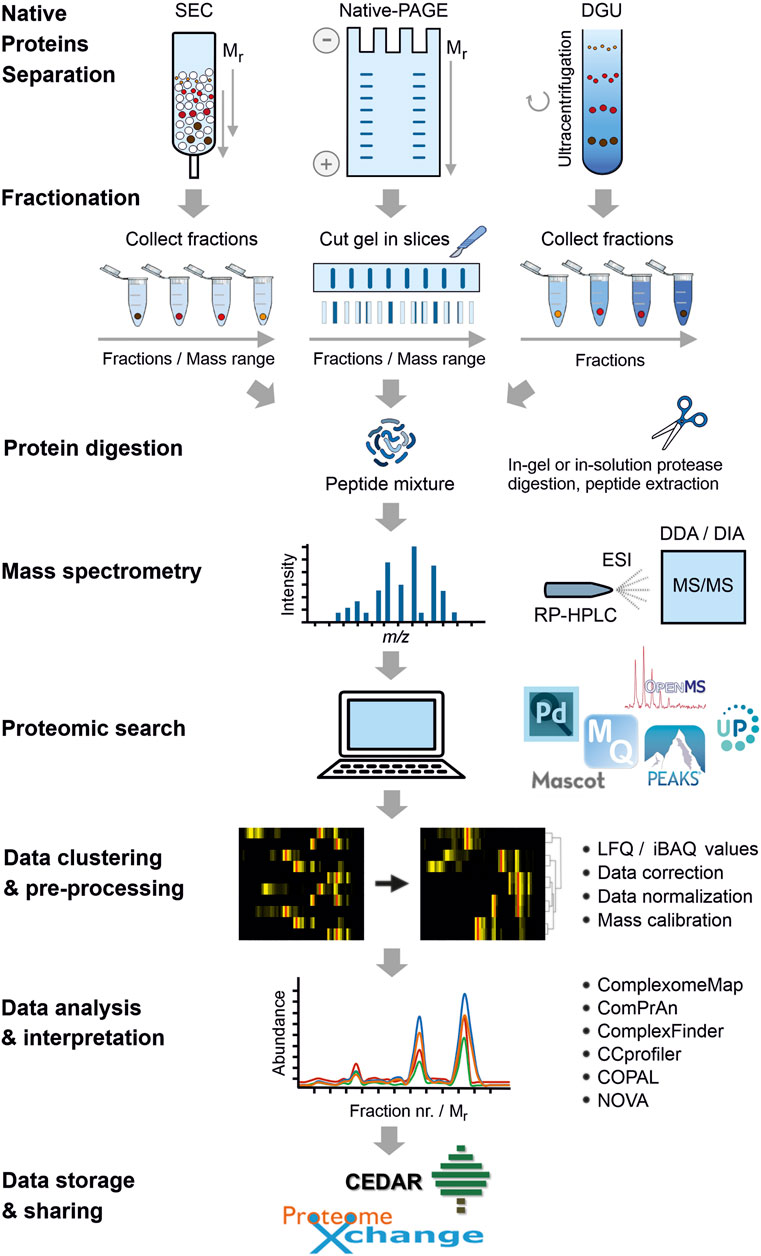
Cabrera-Orefice, A. et al. Frontiers in Cell and Developmental Biology, 2022.
Figure 1. Overall Workflow of Complexome Profiling (CP).
Sample Submission Suggestions
1. Sample Types
Applicable to mammalian cells, tissues, and mitochondria-enriched fractions, with sufficient complex content required.
2. Sample Preparation
Samples should be lysed under cold conditions, avoiding strong detergents or denaturants to preserve the native state of protein complexes. For special handling needs, please consult our technical team in advance.
3. Sample Transportation
All samples should be stored at -80°C and shipped on dry ice to maintain cold-chain conditions and ensure complex stability. For protein extracts, please indicate the buffer composition and protein concentration.
Service Advantages
1. Preservation of Native Complex States
Protein complexes are extracted and separated under non-denaturing conditions to avoid structural disruption and accurately reflect their assembly states in the physiological environment.
2. High-Throughput and High-Sensitivity Detection
Leveraging high-resolution mass spectrometry platforms, the service enables precise identification and quantification of low-abundance proteins within complexes.
3. Visualization of Protein Distribution
By constructing complex migration profiles, the distribution patterns of proteins across fractionation gradients are clearly presented, facilitating the identification of core and accessory components of complexes.
4. Multi-Level Data Interpretation
Integrating interaction networks, functional annotation, and pathway enrichment analysis, this service provides comprehensive insights into functional modules of protein complexes and their dynamic changes under different physiological or pathological conditions.
Applications
1. Mitochondrial Function Studies
Complexome profiling service enables systematic analysis of the assembly states, interaction networks, and dynamic changes of the mitochondrial respiratory chain and supercomplexes under disease or stress conditions.
2. Membrane Protein Complex Analysis
By investigating the composition and function of multiprotein complexes in the plasma membrane or internal membrane systems, this service helps elucidate mechanisms of transmembrane signal transduction and substance transport.
3. Tracking of Protein Assembly Processes
Complexome profiling service allows comparison of migration profiles under different time points or treatment conditions, revealing the sequential order of protein assembly and structural remodeling features.
4. Disease Mechanisms and Biomarker Research
By identifying aberrant complex compositions or disrupted interactions under pathological conditions, this service facilitates the exploration of disease-related protein networks and potential therapeutic targets.
FAQ
Q1: What Is the Difference between Complexome Profiling and Conventional Proteomics?
A1: Conventional proteomics focuses on the expression and function of individual proteins, whereas complexome profiling emphasizes the composition, assembly state, and dynamic changes of multiprotein complexes. It reveals protein cooperation and structural remodeling processes.
Q2: Does the Service Support Comparison Across Multiple Conditions or Time Points?
A2: Yes. Multiple experimental groups can be designed according to research objectives to compare complex assembly and interaction changes under different treatments or time points, making it suitable for mechanistic studies or dynamic process monitoring.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
How to order?







