Comparative Analysis of Shotgun and Targeted Proteomics
-
Discovery stage: Shotgun proteomics combined with DIA for comprehensive data acquisition
-
Validation stage: Custom design of MRM/PRM targets and method development
-
Quantification stage: SIS peptide synthesis for absolute protein quantification
-
Data analysis: Support for network analysis, pathway enrichment, and visual representation
In contemporary proteomics research, the selection of an appropriate mass spectrometry strategy plays a critical role in determining the depth, accuracy, and translational potential of the resulting data and findings. During the initial phase of a study, broad exploratory analyses are often required, for which discovery proteomics—also referred to as the "shotgun approach"—is typically employed. Once candidate targets have been identified, the focus shifts to a verification phase that demands high specificity and reproducibility, where targeted proteomics emerges as the method of choice. This article systematically compares these two strategies in terms of their underlying principles, data characteristics, application scenarios, and relative advantages and limitations. It also presents how MtoZ Biolabs supports researchers in efficiently integrating both approaches to enhance overall research productivity and translational success.
Principle Differences: Exploratory vs. Verificatory
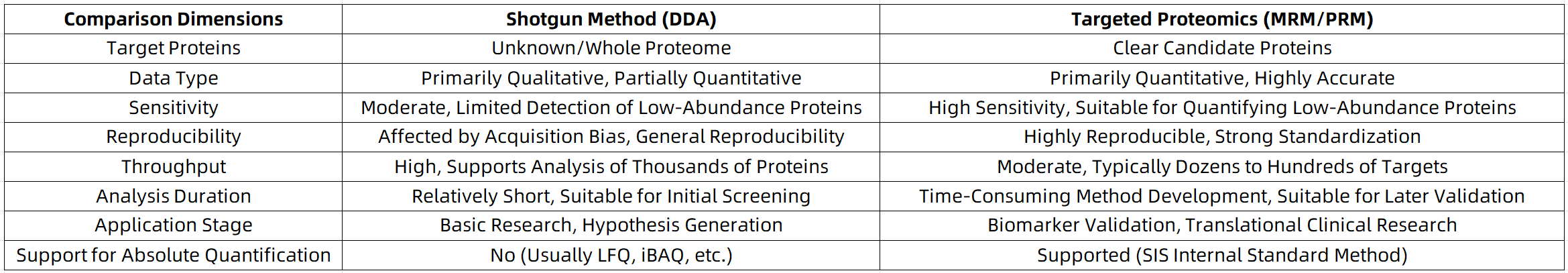
1. Shotgun Proteomics: Large-Scale, Unbiased Discovery
Shotgun proteomics, also known as discovery proteomics, primarily utilizes a data-dependent acquisition (DDA) strategy. Through liquid chromatography-tandem mass spectrometry (LC-MS/MS), the system selectively fragments peptides based on signal intensity and matches them against a protein sequence database. This approach emphasizes proteome coverage and the number of identified proteins, making it well-suited for initial investigations into complex or poorly understood biological mechanisms.
2. Targeted Proteomics: Precision Quantification as a “Sniper”
Targeted proteomics employs methods such as multiple reaction monitoring (MRM) or parallel reaction monitoring (PRM) to detect only predefined peptide targets. The mass spectrometer ignores all non-target information and focuses exclusively on analytes of interest. The resulting data are highly reproducible and quantitatively robust, making this approach ideal for validating candidate biomarkers or disease-associated targets.
Comparative Analysis: Tabular Summary of Strategy Differences
Application Scenarios: Complementary Rather Than Substitutive
1. Typical Applications of Shotgun Proteomics
(1) Large-scale screening of disease-associated differentially expressed proteins (e.g., tumor vs. normal tissue)
(2) Exploration of biological pathways and protein–protein interaction networks
(3) Rapid hypothesis generation and identification of potential biomarkers
2. Typical Applications of Targeted Proteomics
(1) Validation of candidate protein expression in independent sample cohorts
(2) Development of companion diagnostic models to inform clinical decision-making
(3) Absolute quantification of key proteins for pharmacodynamic assessment
MtoZ Biolabs Integrated Solutions: Aligning Technology with Research Objectives
MtoZ Biolabs integrates a range of mass spectrometry strategies, enabling researchers to flexibly select approaches based on specific project needs:
We deliver not just analytical platforms, but a comprehensive, end-to-end service encompassing strategy, analysis, and translational support. Shotgun and targeted proteomics are not mutually exclusive; rather, they form a continuous and integrated research workflow—spanning broad-spectrum discovery, precise targeting, and translational application. The critical factor lies in selecting the appropriate proteomic strategy at each stage of the research process.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







