Antibody-Antigen Complexes Structure Characterization Service
Antibody-Antigen Complexes Structure Characterization Service is a structural biology service based on Cryo-Electron Microscopy (Cryo-EM), focused on resolving the three-dimensional structures of antibody–antigen complexes. It includes precise mapping of binding epitopes, identification of key contact residues, elucidation of steric blocking mechanisms, and analysis of conformational adaptability.
The specific recognition between antibodies and antigens is a fundamental feature of the immune system and forms the scientific basis for monoclonal antibody therapeutics, vaccine design, and diagnostic tool development. Accurate structural elucidation of how antibodies recognize their antigens--particularly the three-dimensional architecture of binding epitopes--is critical for rational antibody design, functional optimization, and mechanistic understanding. Traditional structural methods such as X-ray crystallography face limitations due to requirements for crystal formation and low tolerance for sample heterogeneity. In contrast, Cryo-EM, as a next-generation high-resolution imaging technology, does not require crystallization or regular particle arrangement, making it the leading method for resolving the structures of medium-to-large antibody-antigen complexes. It is especially well-suited for non-crystalline, dynamic, and heterogeneous biological systems.
Leveraging an advanced cryo-electron microscopy platform, MtoZ Biolabs offers the Antibody-Antigen Complexes Structure Characterization Service to analyze a wide range of antigen types—including proteins, glycoproteins, viral particles, and synthetic antigens—as well as antibodies derived from natural sources or through engineering. Our service helps researchers reveal the interaction mechanisms between antibodies and their targets at atomic resolution. Whether you are conducting early-stage antibody screening, functional validation, or in-depth mechanistic studies, we provide robust structural support for your research and development projects.
Analysis Workflow
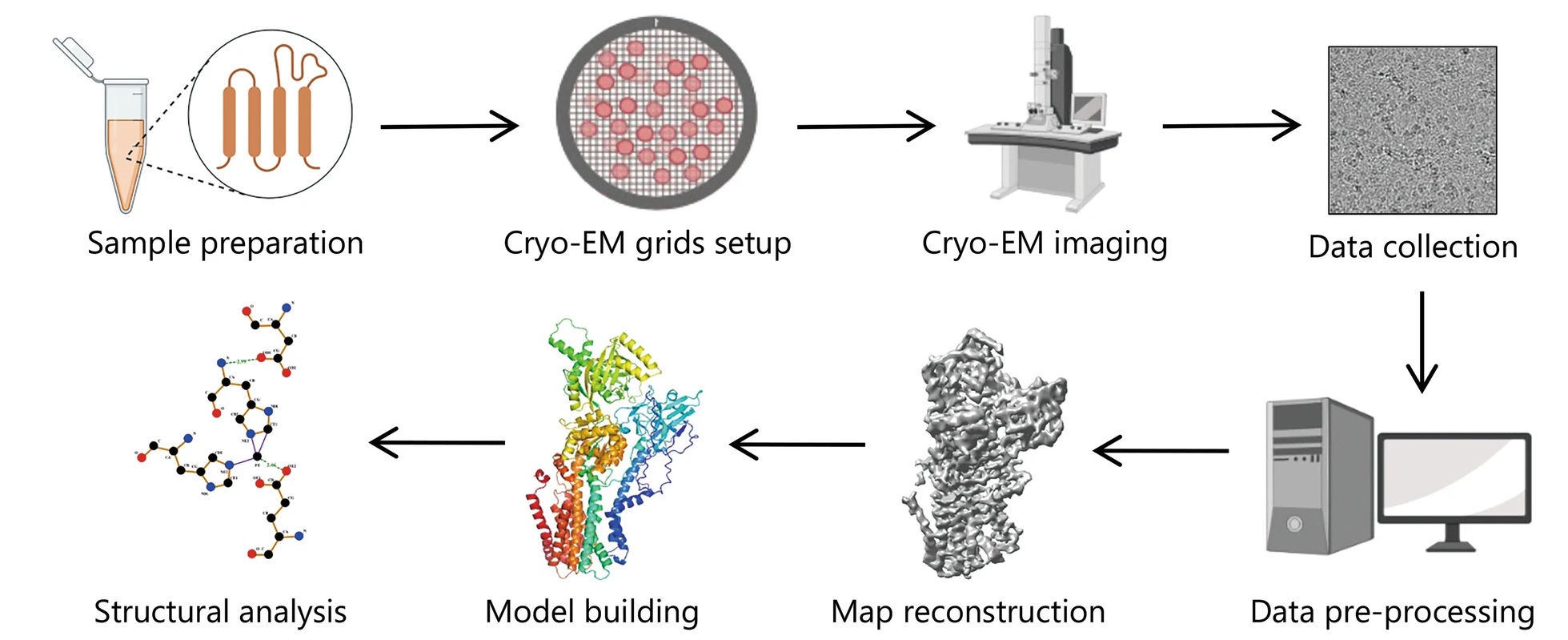
Zhu KF. et al. Military Med Res. 2023.
MtoZ Biolabs offers a comprehensive workflow for Antibody-Antigen Complexes Structure Characterization, supporting everything from antibody screening to mechanistic analysis. The key steps include:
1. Complex Assembly and Preliminary Evaluation
Preparation and purification of the antibody and antigen, followed by complex formation. Techniques such as size-exclusion chromatography (SEC) are used to verify the stability and homogeneity of the complex.
2. Negative-Stain Screening
Negative-stain TEM is used for rapid assessment of complex integrity, particle orientation, and image quality, helping to identify optimal samples for cryo-EM processing.
3. Cryo-EM Sample Preparation and Data Collection
The antibody–antigen complex is vitrified into a layer of amorphous ice, and high-resolution 2D micrographs are collected using a cryogenic transmission electron microscope.
4. 3D Reconstruction and Structural Modeling
Using particle alignment, classification, and reconstruction algorithms, a 3D density map of the complex is generated. Atomic models of the antibody–antigen interaction are built and refined.
5. Structural Interpretation and Epitope Annotation
Detailed analysis of the recognized epitope and key binding residues is performed. Final deliverables include annotated visualizations and a comprehensive structural report.
Applications
Antibody–Antigen Complexes Structure Characterization Service can be applied in a wide range of research and development scenarios, including:
Mechanism of Neutralizing Antibodies Analysis
Analyze whether the antibody blocks functional regions of the antigen (e.g., receptor-binding domains).
Epitope Mapping and Classification
Identify linear and conformational epitopes to support epitope library construction and rational immunogen design.
Affinity Maturation and Antibody Engineering
Design CDR region modifications based on detailed structural insights into the binding interface.
Study on the Mechanisms of Antibody Synergy
Elucidate the cooperative recognition mechanism of bispecific or multispecific antibodies.
Vaccine Antigen Evaluation
Assess the conformational integrity and antibody recognition capacity of candidate vaccine antigens.
Service Advantages
1. Atomic-Level Resolution: High-resolution Cryo-EM enables precise characterization of conformational interactions between antibody CDRs and antigenic epitopes.
2. No Crystallization Required: Suitable for non-crystallizable, heterogeneous, or membrane-associated antigens, including viral particles, glycoproteins, and fusion proteins.
3. Supports Dynamic Conformation Classification and Multi-Antibody Recognition: Enables classification of multiple conformations and identification of overlapping or distinct binding events, aiding the design of synergistic multi-antibody therapeutic strategies.
FAQ
Q. What Types of Antibodies are Supported by this Service?
We support various formats, including full-length IgG, Fab, scFv, and VHH (nanobodies). We can also assist with expression and complex formation if needed.
Q. What Types of Antigens can be Analyzed?
Our service supports a wide range of antigen types, including proteins, glycoproteins, virus-like particles (VLPs), membrane fragments, fusion proteins, and synthetic antigens.
Q. What Sample Requirements should be Met?
We recommend submitting high-purity (>90%) complexes at concentrations above 1 mg/mL. Buffers should avoid glycerol, high salt, or detergents where possible. We can provide sample preparation guidance and complex assembly support.
Case Study
This study used single-particle cryo-electron microscopy (Cryo-EM) technology to perform high-resolution structural characterization of the complex formed by the full-length IgG antibody W328-6H2 and various SARS-CoV-2 Spike protein trimers (including SARS-CoV, the original strain SARS-CoV-2 WT and the Omicron BA.1 variant). The results showed that W328-6H2 IgG can mediate inter-spike cross-linking through its bivalent Fab region and induce the formation of higher-order aggregates between Spikes, which appear as "head-to-head" symmetrical dimers in the SARS-CoV and WT backgrounds, and exhibit conformational shifts in Omicron BA.1. This antibody-induced aggregation pattern is considered to be a key mechanism for its enhanced neutralizing activity and also provides a structural basis for designing broad-spectrum antibodies with cross-variant neutralizing capabilities.
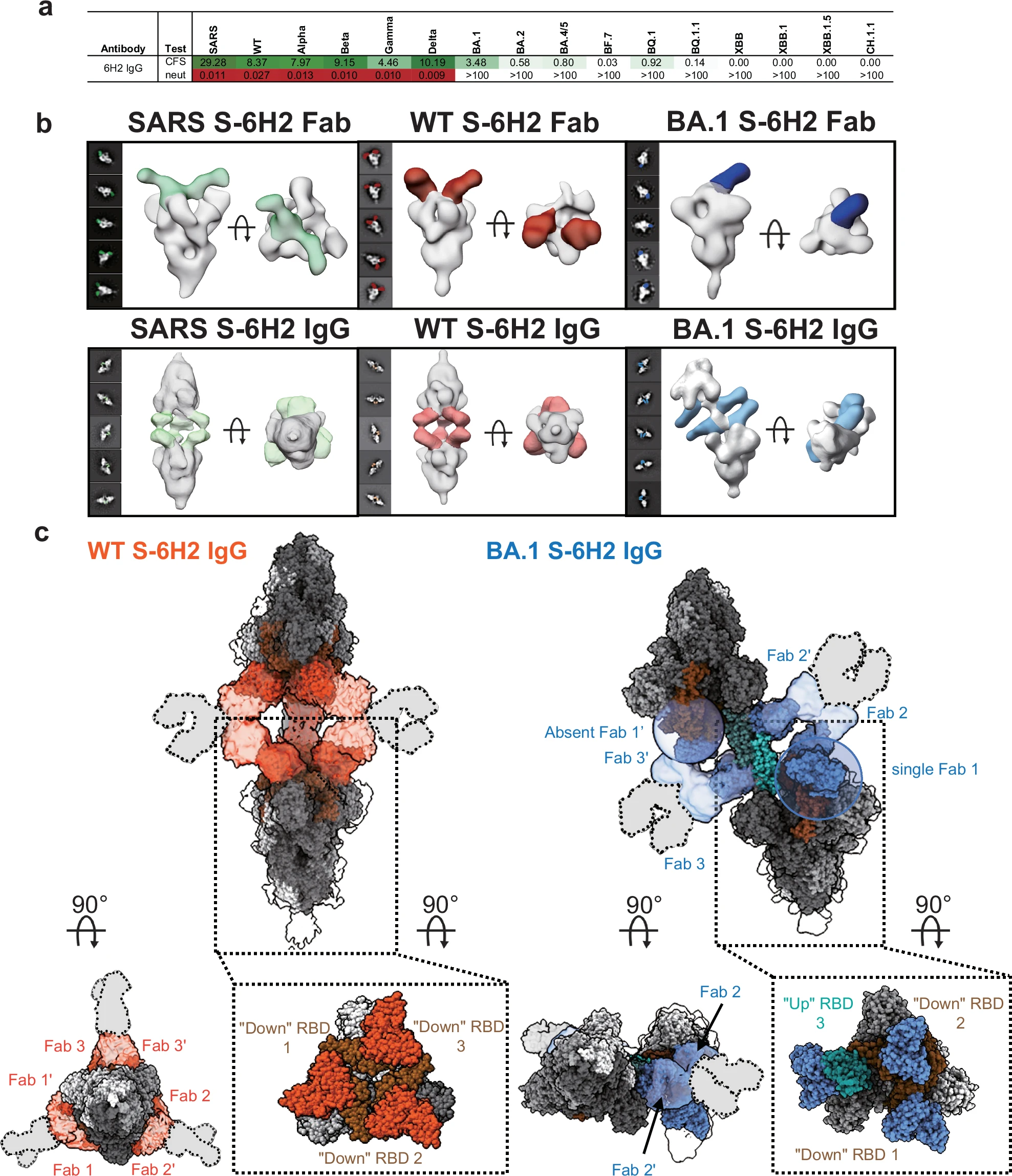
Nan X. et al. Nat Commun. 2024.
How to order?







