Advantages and Limitations of Targeted Proteomics
-
MRM, utilizing a triple quadrupole mass spectrometer, offers excellent selectivity and is well-suited for high-throughput biological sample analysis.
-
PRM, based on high-resolution mass spectrometry, acquires complete MS/MS spectra, effectively minimizing ion interference.
-
The use of predefined peptide targets and acquisition windows significantly reduces technical variability.
-
Incorporation of stable isotope-labeled peptides (SIS) allows for highly accurate absolute quantification, supporting the generation of data in compliance with clinical regulatory standards.
-
Quantitative validation of biomarkers;
-
Development of companion diagnostic approaches;
-
Pharmacokinetic/pharmacodynamic (PK/PD) studies of therapeutic agents.
-
Screening of signature peptides;
-
Optimization of CE/RT parameters;
-
Incorporation of internal standards and method validation;
-
Instrument calibration and establishment of quality control protocols.
-
MRM throughput: 50–200 targets (in high-sensitivity mode)
-
PRM throughput: 100–300 targets (constrained by retention time window)
-
Target Selection: Identification and recommendation of optimal signature peptides based on databases, published literature, or DDA (Data-Dependent Acquisition) results.
-
SIS Internal Standard Synthesis: In-house synthesis platform enabling the customization of high-purity, stable isotope-labeled peptides.
-
Method Development and Optimization: Established standard workflows for MRM (Multiple Reaction Monitoring) and PRM (Parallel Reaction Monitoring), which can be rapidly implemented.
-
Data Analysis and Visualization: Comprehensive support for Skyline-based data analysis, functional annotation, and the construction of biological pathway diagrams.
As life science research transitions from “discovery” to “validation,” greater demands have been placed on the accuracy, reproducibility, and quantitative capabilities of proteomic data. Targeted Proteomics has therefore emerged as a critical bridge between basic research and clinical translation. In particular, technologies such as Multiple Reaction Monitoring (MRM) and Parallel Reaction Monitoring (PRM), known for their high controllability and reproducibility, have been widely employed in areas including biomarker validation, companion diagnostics, and investigations of drug mechanisms. Despite its precision, this approach also entails certain limitations. This article provides a comprehensive overview of the principal advantages and limitations of Targeted Proteomics and highlights how MtoZ Biolabs has optimized performance in this domain.
Core Advantages of Targeted Proteomics
1. High Quantitative Sensitivity and Specificity
Targeted Proteomics enhances signal intensity and reduces background noise through repeated monitoring of specific peptides and ion pairs, making it particularly effective for the detection of low-abundance proteins.
The Targeted Proteomics platform at MtoZ Biolabs can theoretically achieve detection sensitivity at the picogram-per-milliliter (pg/mL) level and is compatible with diverse sample types, including cerebrospinal fluid and tissue homogenates.
2. Excellent Reproducibility and Methodological Robustness
Targeted mass spectrometry exhibits strong reproducibility and inter-batch consistency, making it ideal for multicenter and long-term studies.
3. Strong Suitability for Clinical Translation
Compared to discovery-based methods such as Data-Dependent Acquisition (DDA), Targeted Proteomics is better suited for downstream validation and quantitative analysis, and can be effectively applied in:
Major Limitations of Targeted Proteomics
1. Dependence on Prior Knowledge and Limited Novel Discovery Capability
Targeted approaches rely on known protein sequences or literature-derived data, and are therefore unsuitable for identifying novel proteins, post-translational modifications, or previously unknown biological pathways. As a result, initial studies typically require discovery-based strategies such as DDA or DIA to screen for candidate targets prior to entering the targeted validation phase.
2. Lengthy Development Process and High Technical Demands
Developing high-quality targeted proteomics methods involves several critical steps, including:
These processes demand substantial expertise from technical personnel and a high degree of stability in the mass spectrometry platform. MtoZ Biolabs offers end-to-end method development services, with standard projects deliverable within 7–10 business days, thereby reducing project timelines.
3. Limited Throughput, Restricting Simultaneous Quantification of Large Protein Sets
Currently, targeted techniques support:
For projects requiring quantification of more than 500 proteins, a combined DIA and DIA-PRM strategy is recommended to achieve broader coverage.
Targeted Proteomics vs. Other Mass Spectrometry Strategies
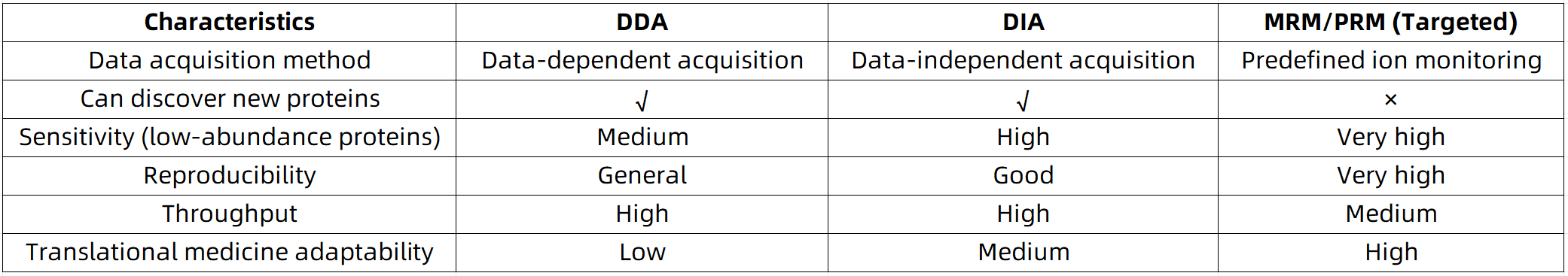
MtoZ Biolabs: One-Stop Service from “Target Design” to “Data Generation”
MtoZ Biolabs has developed an integrated and systematic workflow for targeted proteomics:
While targeted proteomics is not a universal solution, it remains one of the most reliable approaches for quantitative studies that require high precision, reproducibility, and standardization. Researchers are encouraged to strategically integrate both targeted and untargeted proteomics approaches, depending on the research phase, the number of target proteins, and the complexity of the samples, to enhance overall research efficiency. MtoZ Biolabs is committed to providing a high-performance and highly consistent technical platform for quantitative proteomics, empowering researchers to effectively translate their scientific findings into practical applications.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







