10x RNA Seq Service
- Standard Transcriptome Analysis (e.g., tumor and immune cells): 20,000–50,000 reads/cell, suitable for most biological studies.
- Low-abundance Transcript Detection (e.g., neurons, stem cells): 50,000–100,000 reads/cell, enhances sensitivity and reduces dropout effects.
- Advanced Applications (e.g., whole-transcriptome analysis): 100,000+ reads/cell, ideal for detecting lncRNAs or splicing variations.
10x RNA Seq Service utilizes the 10x Genomics Chromium platform for single-cell transcriptome sequencing, enabling the analysis of gene expression at the single-cell level to reveal cellular heterogeneity and molecular characteristics. 10x RNA Seq is based on microfluidic chip technology and employs Gel Beads-in-Emulsion (GEMs) to achieve single-cell partitioning, while incorporating Unique Molecular Identifiers (UMIs) to label mRNA, ensuring quantification of gene expression at the single-cell level.
10x RNA Seq Service enables the precise quantification of transcript expression across thousands to hundreds of thousands of single cells, allowing for the identification of cell types, functional states, and developmental trajectories. 10x RNA Seq technology is widely applied in tumor microenvironment studies, immunology, neuroscience, stem cell research, and disease mechanism investigations, providing robust data support for precision medicine and biological research.
Utilizing the 10x Genomics Chromium system in combination with the Illumina platform, MtoZ Biolabs offers 10x RNA Seq Service including cell sorting, library preparation, sequencing, and data analysis, empowering researchers to precisely decode gene regulatory networks at the single-cell resolution.
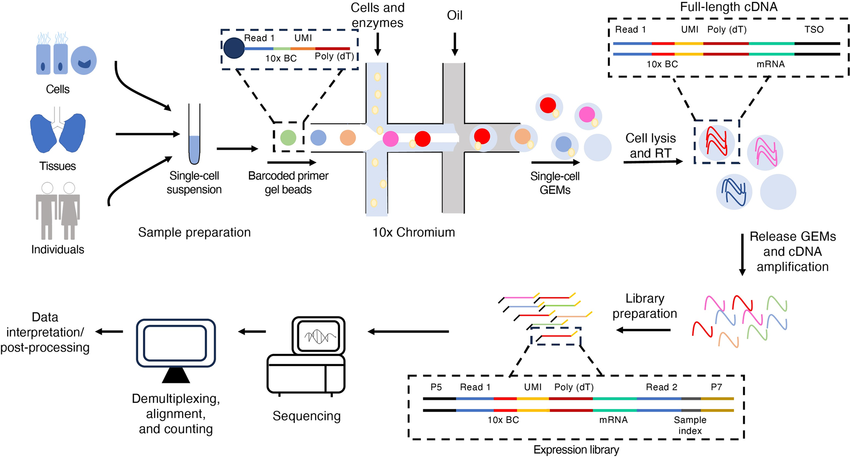
Swaminath S. Russell A B. PLoS Pathogens. 2024.
Analysis Workflow
1. Sample Preparation
Prepare fresh tissue or cell suspension, and perform single-cell sorting into the 10x microfluidic chip.
2. 10x Genomics Single-Cell Library Construction
Generate single-cell GEMs. Perform reverse transcription (RT) and PCR amplification of RNA to construct the sequencing library.
3. High-Throughput Sequencing
Utilize the Illumina platform for high-throughput sequencing to obtain single-cell transcriptomic data.
4. Data Analysis
Quality control and filtering (removal of low-quality cells and background noise).
Cell clustering (visualization using PCA, t-SNE, and UMAP).
Differential gene expression analysis and cell type identification.
Applications
Common applications of 10x RNA Seq Service:
Tumor Microenvironment Research
Uncover the heterogeneity of immune cells, fibroblasts, vascular cells, and other cells within the tumor microenvironment (TME). Analyze tumor progression, drug resistance, and immune evasion mechanisms.
Immunology and Infection Research
Characterize immune cell differentiation trajectories, functional states, and disease-associated changes.
Neuroscience
Study the cell type spectrum of the nervous system and explore the molecular mechanisms of neurodevelopmental and degenerative diseases.
Stem Cell and Developmental Biology
Trace stem cell differentiation pathways and dissect key gene regulatory networks involved in tissue development.
Precision Medicine
Identify disease-specific single-cell transcriptomic signatures to provide molecular targets for personalized therapeutic strategies.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced 10x RNA Seq platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all 10x RNA Seq data, providing clients with a comprehensive data report.
4. High-throughput Single-cell Sequencing: Capable of analyzing tens of thousands of cells in a single run, enabling large-scale studies of cellular heterogeneity.
FAQ
Q. How to identify and remove doublets in 10x RNA Seq data?
Doublets occur when two or more cells are mistakenly encapsulated within the same droplet (GEM), leading to false-positive co-expression of genes, which can affect downstream analysis. The following methods can help detect and remove doublets:
1. Computational Methods: Use tools such as DoubletFinder (based on nearest neighbor clustering), Scrublet (based on UMI count distribution), or scDblFinder to detect and filter out cell populations with abnormally high UMI/gene counts.
2. Experimental Optimization: Reduce the input cell concentration (recommended ≤1000 cells/µL) to minimize the occurrence of doublets.
3. Dimensionality Reduction and Clustering Inspection: In UMAP/t-SNE clustering, if certain cell clusters are located between two known cell types, they may be doublets and require further validation.
Q. How to select the sequencing depth (reads per cell) while balancing cost and data quality?
Sequencing depth (reads per cell) determines the detection capability of mRNA in each single cell and should be adjusted according to the study requirements:
If the budget is limited, you can first perform low-depth sequencing (10,000 reads/cell), evaluate data saturation through data dilution experiments, and then decide on the final sequencing depth.
Case Study
This study utilized 10x RNA Seq to analyze the dynamic gene expression changes during chemical reprogramming. The results revealed that cells undergo a transcriptional regulatory pattern of early embryonic-like programs during reprogramming and gradually acquire pluripotency. The study identified key transcription factor regulatory networks and uncovered the influence of specific genes on reprogramming efficiency. These findings provide new insights into gene-free stem cell reprogramming, offering potential new strategies for regenerative medicine and cell therapy.
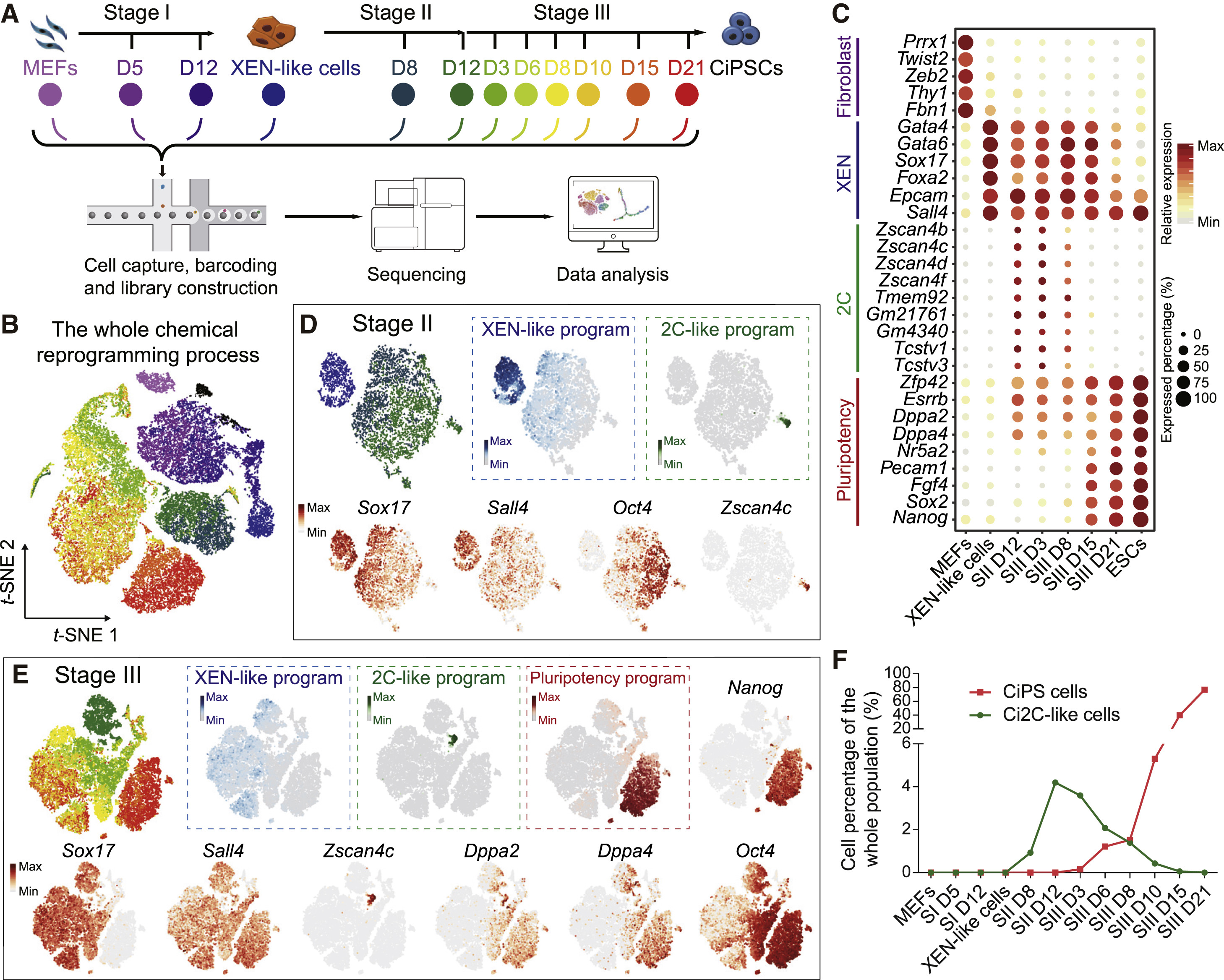
Ting Z. et al. Cell Stem Cell. 2018.
How to order?







