10x Genomics Illumina Sequencing Service
10x Genomics Illumina Sequencing Service integrates 10x Genomics' microfluidic library preparation technology with Illumina’s high-throughput sequencing, delivering high-resolution data across single-cell transcriptomics, epigenomics, and long-fragment DNA genome sequencing. In this service, 10x Genomics is responsible for single cell capture, long-fragment DNA reconstruction, unique molecular identifiers labeling, and efficient library preparation, ensuring high-quality samples for sequencing. Illumina sequencing then processes the 10x Genomics-prepared libraries with high-throughput, high-accuracy short-read sequencing, generating high-resolution single-cell genomic data. Leveraging the 10x Genomics Chromium system and the Illumina platform, MtoZ Biolabs offers 10x Genomics Illumina Sequencing Service to support precision medicine, biomedical research, and multi-omics studies.
Technical Principles
10x Genomics technology utilizes microfluidics to encapsulate single cells or long-fragment DNA into gel beads-in-emulsion (GEMs). It employs unique molecular identifiers (UMIs) to label mRNA, DNA, or chromatin, followed by Illumina high-throughput sequencing. This technology is characterized by high resolution, low bias, and high throughput, enabling single-cell-level analysis of transcriptomics, genomics, and epigenetics.
Analysis Workflow
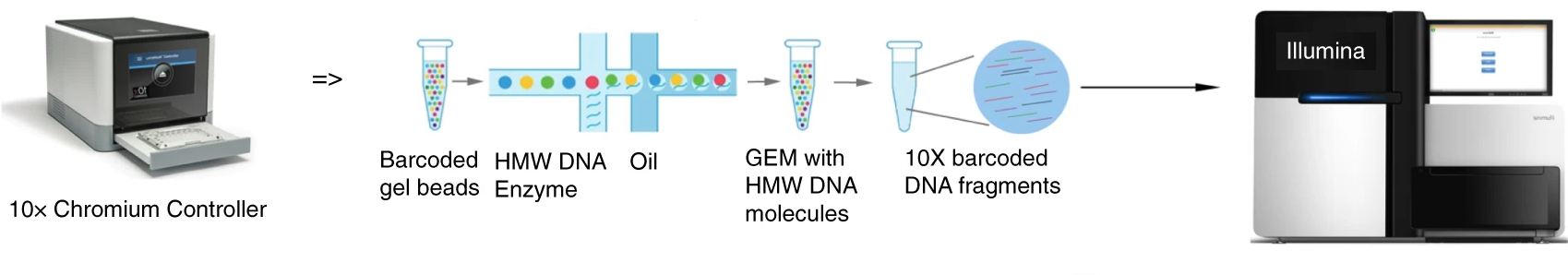
Sun H. et al. Nature Communications. 2018.
Core steps of 10x Genomics Illumina Sequencing Service:
1. Sample Preparation
Preparation of single-cell suspensions, ultra-high molecular weight (HMW) DNA, T-cell/B-cell samples, or fragmented chromatin.
2. 10x Genomics Library Construction
Single-cell mRNA capture (10x Chromium scRNA-Seq); Long-fragment DNA barcoding (10x Linked-Reads WGS); T/B cell receptor sequencing (10x V(D)J-Seq).
3. Illumina High-Throughput Sequencing
High-depth short-read sequencing to ensure data accuracy.
4. Data Analysis
Cell clustering, gene expression quantification, haplotype analysis, structural variation (SV) detection, etc.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Leveraging the 10x Genomics Chromium system and Illumina platform, MtoZ Biolabs provides 10x Genomics Illumina Sequencing Service covering various research areas, including scRNA-Seq, scATAC-Seq, and scVDJ-Seq.
Applications
1. Single-Cell RNA Sequencing (10x scRNA-Seq)
Analyzes single-cell heterogeneity, identifying cell types and developmental trajectories.
Applied to cancer microenvironment studies, immune system research, and neuroscience.
2. Long-Fragment DNA Sequencing (10x Linked-Reads WGS)
Resolves complex genomes, including high-GC content and repetitive regions.
Detects structural variations (SVs), applicable to cancer genomics and rare disease research.
Enables haplotype phasing, reconstructing parental haplotype information.
3. Single-Cell ATAC Sequencing (10x scATAC-Seq)
Investigates chromatin accessibility and transcription factor regulatory networks.
Integrates with 10x RNA-Seq data for multi-omics analysis.
4. Immune Repertoire Sequencing (10x V(D)J-Seq)
Profiles T-cell and B-cell receptors to study adaptive immune responses.
Analyzes tumor immune microenvironments and identifies specific TCR/BCR sequences.
5. Spatial Transcriptomics (10x Visium Spatial Transcriptomics)
Maps gene expression spatially within tissue sections.
FAQ
Q. How to Identify and Remove Low-Quality Cells to Improve Data Accuracy?
Low-quality cells are typically characterized by low gene expression levels, low UMI counts, and a high proportion of mitochondrial genes. The following methods can be used for quality control:
1. Gene Count Filtering: Set a threshold for the number of genes expressed per cell, typically between 200 and 5000. Remove outliers with too few genes (empty droplets) or too many genes (doublets).
2. UMI Counts Filtering: Exclude cells with extremely low UMI counts (typically below 1000 UMIs) to reduce noise in the dataset.
3. Mitochondrial Gene Proportion: Remove cells where mitochondrial gene expression exceeds 10-15%, to reduce the effects of cellular stress or apoptosis.
4. Data Visualization Inspection: Use t-SNE or UMAP for dimensionality reduction to identify abnormal cell populations and refine filtering criteria.
Case Study
This study utilized 10x Genomics Illumina sequencing to analyze the heterogeneity of skeletal muscle cells in a dystrophic mouse model. The findings revealed that as the disease severity increased, the cellular composition of muscle tissue underwent significant changes, characterized by an increase in fibroblasts and immune cells, while the function of muscle stem cells (satellite cells) was impaired. Further analysis identified key changes in gene expression patterns, particularly in pathways related to inflammation, fibrosis, and muscle regeneration. Ultimately, the study identified several potential therapeutic targets, providing new insights into the molecular mechanisms of muscular dystrophy and the development of targeted therapies.
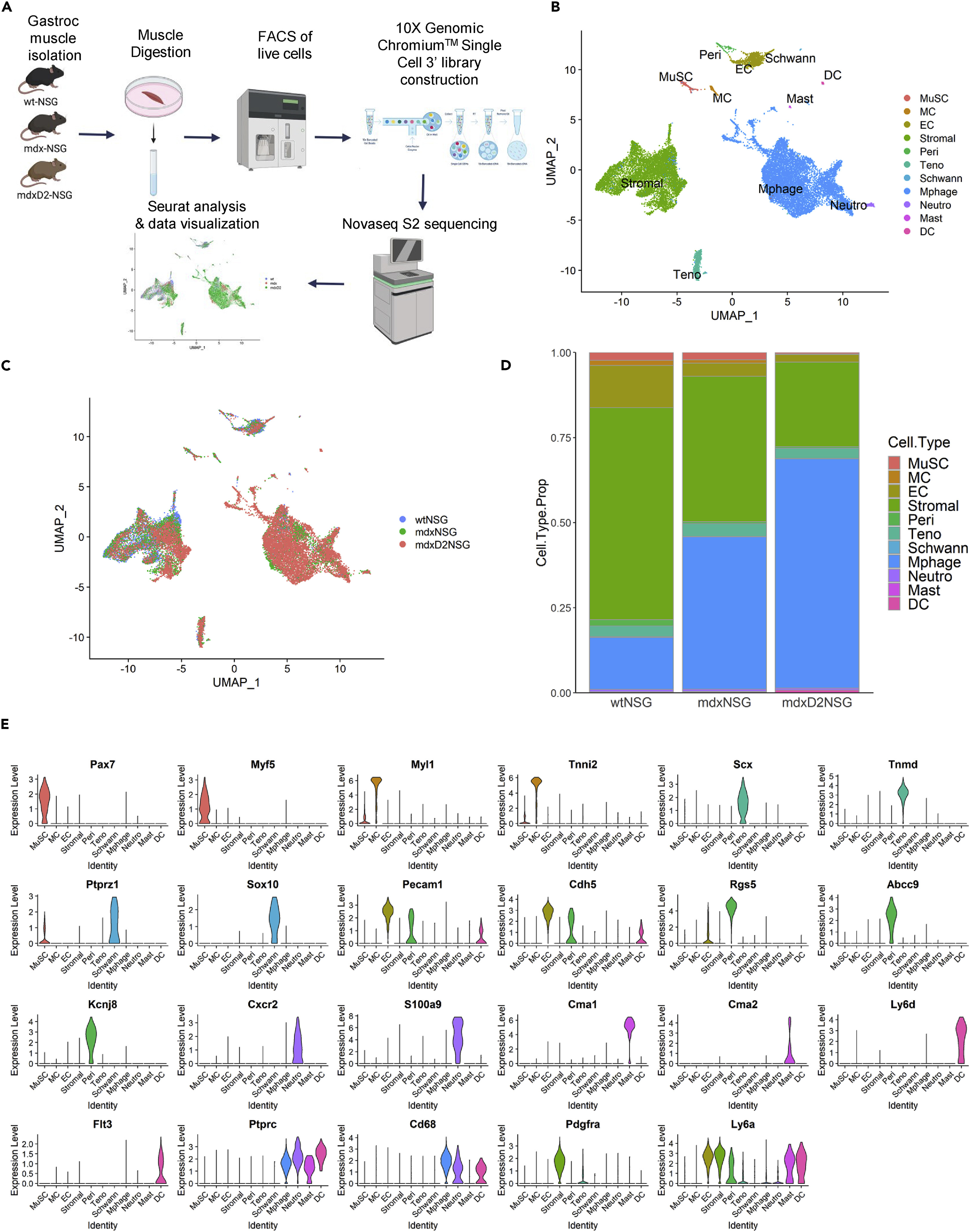
Saleh K K. et al. iScience. 2022.
How to order?







