What Tools Are Used for Protein Sequence Analysis? A Summary of Common Methods and Techniques
-
InterProScan: https://www.ebi.ac.uk/interpro/
-
Pfam (Protein Families Database): http://pfam.xfam.org
-
BLASTp: https://blast.ncbi.nlm.nih.gov
-
Clustal Omega (multiple sequence alignment): https://www.ebi.ac.uk/Tools/msa/clustalo/
-
TMHMM (Transmembrane domain prediction): https://services.healthtech.dtu.dk/service.php?TMHMM-2.0
-
SignalP (Signal peptide prediction): https://services.healthtech.dtu.dk/service.php?SignalP
-
DeepLoc: https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0
-
WoLF PSORT: https://wolfpsort.hgc.jp
-
NetPhos (phosphorylation site prediction): https://services.healthtech.dtu.dk/service.php?NetPhos-3.1
-
GPS (comprehensive PTM prediction): http://gps.biocuckoo.cn
-
BepiPred 2.0 (B-cell epitopes): https://services.healthtech.dtu.dk/service.php?BepiPred-2.0
-
IEDB Analysis Resource (comprehensive T/B cell prediction): https://www.iedb.org
-
AlphaFold DB: https://alphafold.ebi.ac.uk
-
I-TASSER: https://zhanggroup.org/I-TASSER/
-
Functional annotation of novel proteins or those from non-model species
-
Precise identification of antibody light/heavy chains and CDR regions
-
Integration of PTM site prediction with mass spectrometry validation
-
Epitope screening enhanced by AlphaFold-based structural visualization
-
Automated batch annotation and generation of graphical analysis reports
In fields such as proteomics, structural biology, functional annotation, and drug discovery, protein sequence analysis frequently serves as a foundational step in research. This type of analysis not only determines the primary structure of proteins but also supports the prediction of molecular features such as functional domains, signal peptides, transmembrane regions, post-translational modification sites, and linear antigenic epitopes. It constitutes a critical component in identifying potential targets and designing candidate molecules.
What are the most commonly used tools for protein sequence analysis, and what specific tasks are they best suited for? This article presents a systematic overview of seven key tasks involved in protein sequence analysis, along with representative tools for each. Practical recommendations are also provided to help researchers enhance their analytical efficiency and streamline experimental workflows.
Core Tasks in Protein Sequence Analysis
In practice, protein sequence analysis typically encompasses the following objectives:
1. Initial functional annotation of proteins
2. Identification of structural domains and conserved motifs
3. Prediction of subcellular localization (e.g., membrane-associated proteins)
4. Prediction of post-translational modification sites (e.g., phosphorylation, acetylation)
5. Detection of signal peptides and transmembrane regions
6. Epitope prediction (for vaccine or antibody development)
7. Comparative analysis of homologous proteins or protein families
Seven Common Categories of Protein Sequence Analysis Tools and Recommendations for Use
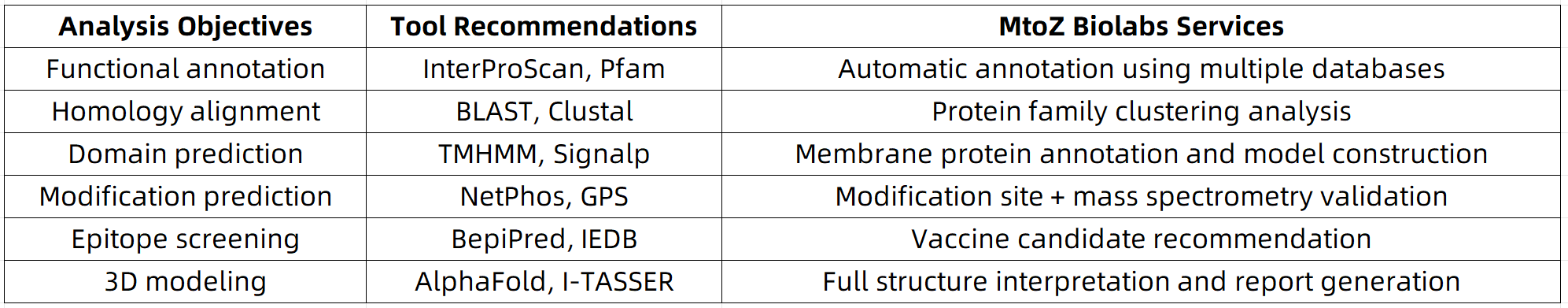
1. Protein Function Annotation Tools: InterPro & Pfam
(1) Function: Identify protein domains, functional families, enzyme classifications, and Gene Ontology (GO) terms
(2) Recommended tools:
(3) Recommendations for use:
InterProScan integrates multiple databases (Pfam, PRINTS, SMART, etc.) and returns comprehensive annotation results from a single sequence input, making it especially suitable for preliminary functional characterization of novel proteins.
2. Sequence Homology Tools: BLAST & Clustal Omega
(1) Function: Compare sequences against known databases to identify homologous proteins and infer functional or evolutionary relationships
(2) Recommended tools:
(3) Recommendations for use:
BLAST is ideal for rapid homology searches of individual sequences, while Clustal Omega is better suited for aligning multiple sequences, enabling phylogenetic analysis or identification of conserved regions.
3. Transmembrane Domain and Signal Peptide Prediction Tools: TMHMM & SignalP
(1) Function: Predict the presence of transmembrane domains or signal peptides to assess whether a protein is membrane-bound or secreted
(2) Recommended tools:
(3) Recommendations for use:
Because membrane proteins often contain hydrophobic transmembrane regions, prediction accuracy may vary. It is advisable to perform cross-validation using multiple tools for more reliable results.
4. Subcellular Localization Prediction Tools: DeepLoc / WoLF PSORT
(1) Function: Predict the primary subcellular localization of proteins (e.g., nucleus, mitochondria, secretory pathway)
(2) Recommended Tools:
(3) Usage Suggestions:
Tool selection should align with the biological source of the sample (e.g., human, plant, fungal). DeepLoc is optimized for eukaryotic animal proteins, while WoLF PSORT demonstrates broader applicability across diverse taxa.
5. Post-Translational Modification Prediction Tools: NetPhos / GPS
(1) Function: Predict potential sites of post-translational modifications, such as phosphorylation, acetylation, and ubiquitination
(2) Recommended Tools:
(3) Usage Suggestions:
As computational predictions indicate only putative modification sites, experimental validation (e.g., PTM enrichment and LC-MS/MS analysis) is strongly recommended to confirm biological relevance.
6. Antigen Epitope Prediction Tools: BepiPred / IEDB Toolkit
(1) Function: Predict B-cell and T-cell antigen epitopes, widely used in vaccine development and antibody therapeutics
(2) Recommended Tools:
(3) Usage Suggestions:
Combining linear epitope prediction with structural accessibility and sequence conservation analysis can improve the interpretability of results. Structural modeling tools (e.g., AlphaFold) may aid in epitope selection.
7. Protein Tertiary Structure Prediction Tools: AlphaFold & I-TASSER
(1) Function: Predict 3D structure from the primary amino acid sequence to facilitate functional studies
(2) Recommended Tools:
(3) Usage Suggestions:
AlphaFold is best suited for predicting structures of full-length proteins, while I-TASSER is more appropriate for modeling protein fragments. Predicted structures can be visualized using molecular visualization tools such as PyMOL or UCSF Chimera.
MtoZ Biolabs: One-Stop Protein Sequence Analysis Service Platform
While the aforementioned tools enable diverse protein sequence analyses, researchers often encounter several practical challenges, including heterogeneous data sources, complexity in integrating results, and labor-intensive batch annotations. Furthermore, it remains difficult to systematically correlate prediction results with experimental proteomics, structural genomics, or post-translational modification data. Leveraging years of expertise in proteomics and structural biology services, MtoZ Biolabs has developed a high-throughput, automated platform that integrates leading sequence analysis tools and bioinformatics algorithms. This one-stop platform supports:
We aim to support research teams and pharmaceutical enterprises by delivering reliable, interpretable, and traceable data for protein function analysis, thereby facilitating the discovery of valuable scientific insights.
For inquiries regarding protein sequence analysis, functional prediction, or proteomics research, please contact the technical support team at MtoZ Biolabs. Complimentary technical consultations and assistance with project design are available upon request.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







