What Is Peptidomics? Overview of Its Workflow and Principles
-
Capture endogenous signaling peptides that reflect the in vivo protein degradation profile.
-
Characterize their sequences, origins, and variations in abundance.
-
Associate peptide functions with disease states, pharmacological responses, or physiological conditions.
-
Support for plasma, urine, cerebrospinal fluid, and cell culture supernatants
-
One-stop DIA quantification and PRM validation workflow
-
Integrated pathway analysis combining peptidomics with proteomics and metabolomics
-
Delivery of professional visualization reports and SCI publication-ready figures
Proteomics has long been recognized as a pivotal discipline for deciphering cellular functions and elucidating disease mechanisms. In recent years, however, growing attention has been directed toward the degradation products of proteins, endogenous peptides, which harbor valuable physiological and pathological information. Peptidomics has thus emerged as a crucial bridge linking protein expression to functional regulation.
What Is Peptidomics?
Peptidomics is the systematic investigation of naturally occurring low-molecular-weight peptides in biological specimens. These peptides are typically generated through specific protease-mediated cleavage of proteins under physiological conditions, and they often exhibit well-defined biological activities or signaling regulatory roles.
Distinction from Proteomics
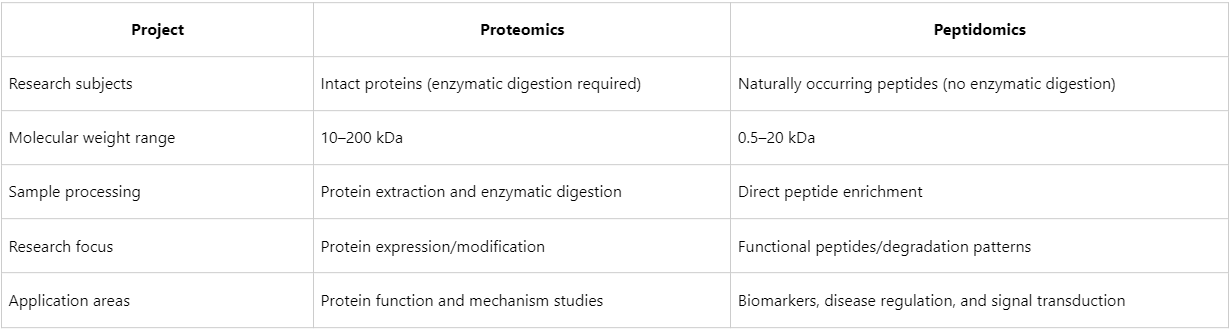
Research Principles of Peptidomics
At its core, peptidomics involves the direct detection of endogenous peptides present in samples using high-resolution mass spectrometry, followed by their identification, quantification, and functional characterization. The fundamental framework of peptidomics research can be outlined in three sequential stages:
Given that these peptides frequently act as enzymatic products, regulatory mediators, or potential immune epitopes, their study offers substantial translational potential for biomarker discovery, drug development, and immunotherapeutic applications.
Standardized Experimental Workflow in Peptidomics
1. Sample Collection and Processing
(1) Common sample types include plasma, urine, cerebrospinal fluid, and cell culture supernatants.
(2) Protease inhibitors are added to prevent post-collection peptide degradation.
2. Peptide Enrichment and Pretreatment
(1) Representative methods include C18 solid-phase extraction (SPE), ultrafiltration, and hydrophilic interaction liquid chromatography (HILIC).
(2) The aim is to remove high-abundance proteins and salts while concentrating target peptides.
3. Mass Spectrometry Analysis
(1) LC-MS/MS platforms (e.g., Orbitrap or Q-TOF) are employed for peptide separation and high-precision qualitative analysis.
(2) Acquisition modes include DDA (data-dependent acquisition) and DIA (data-independent acquisition).
4. Bioinformatics Analysis
(1) Identify peptide sequences and map them to their corresponding protein origins.
(2) Conduct differential expression analysis, clustering, and pathway enrichment.
(3) Perform functional predictions (e.g., SignalP, PeptideRanker) and visualize results.
5. Targeted Validation (Optional)
(1) Validate putative peptides using PRM or SRM approaches.
(2) Complement with ELISA or immunoassays to enhance clinical relevance.
Applications of Peptidomics
1. Biomarker Discovery
Endogenous peptides, regulated by proteolytic processing, are closely linked to disease pathophysiology, making them valuable for early detection, subtype classification, and prognosis monitoring.
2. Elucidating Drug Mechanisms and Targets
Pharmacological interventions often alter protein degradation patterns; peptidomics can uncover pathway activation states and target-specific responses.
3. Immune Epitope Prediction and Vaccine Design
As the primary source of antigenic peptides presented by MHC molecules, peptides analyzed through peptidomics, particularly when integrated with immunomics, can inform the design of precision vaccines.
4. Investigating Mechanisms of Neurological and Metabolic Disorders
Examples include β-amyloid-derived peptides in Alzheimer’s disease and insulin-signaling regulatory peptides in the urine of diabetic patients, both of which represent active research frontiers.
Peptidomics Platform at MtoZ Biolabs
With extensive experience in life sciences research services, MtoZ Biolabs has developed a versatile peptidomics platform compatible with multiple sample types, addressing needs from basic research to clinical translation:
Peptides serve both as molecular traces of protein degradation and as codes of regulatory signaling. As an emerging and rapidly evolving discipline, peptidomics is steadily unveiling the systematic insights and clinical potential embedded within these molecular fragments. MtoZ Biolabs seeks to collaborate with researchers worldwide to advance this promising field and accelerate the translation of scientific discoveries into practical applications.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







