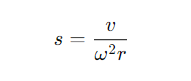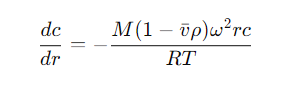Ultracentrifugation for Protein Molecular Weight Determination: Methodologies, Strategies, and the Present Scenario
Protein molecular weight determination is a critical method for studying the structure, function, and biological properties of proteins. Ultracentrifugation, based on centrifugal forces and solution dynamics, plays a pivotal role in determining protein molecular weight, aggregation states, and interactions. Unlike SDS-PAGE, mass spectrometry, and gel filtration, ultracentrifugation directly measures the true molecular weight of proteins in solution, without the need for external calibration standards. This article explores the principles, experimental strategies, data analysis methods, and recent applications of ultracentrifugation in protein research.
Basic Principle of Ultracentrifugation
Ultracentrifugation utilizes high-speed rotation to generate centrifugal forces, causing proteins to sediment in solution. Their molecular weight is calculated based on the sedimentation rate or equilibrium. The common approaches are Sedimentation Velocity (SV) and Sedimentation Equilibrium (SE). Sedimentation Velocity analysis observes how proteins sediment over time under centrifugal forces. This method is ideal for molecular weight determination, homogeneity assessment, and analyzing protein aggregation. Sedimentation Equilibrium focuses on the protein concentration distribution when equilibrium is reached. It provides high-precision molecular weight measurements, detecting protein-ligand binding and oligomer analysis.
1. Sedimentation Velocity (SV)
In SV experiments, the sedimentation coefficient (s) of proteins is measured under high-speed centrifugation, from which molecular weight is calculated. The formula for the sedimentation coefficient is:
Figure 1
s: Sedimentation coefficient (Svedberg, S)
v: Sedimentation velocity
ω: Angular velocity (rad/s)
r: Radius from the particle to the axis of rotation
(1) Data Analysis
①The Lamm equation, which models sedimentation and diffusion, is used to fit the sedimentation curve and calculate molecular weight.
②Density gradient centrifugation differentiates proteins of various molecular weights.
(2) Applications
①Protein aggregation state analysis: Can distinguish between monomers, dimers, and higher-order oligomers.
②Protein complex composition measurement: Useful for studying protein-protein and protein-nucleic acid interactions.
2. Sedimentation Equilibrium (SE)
SE is a non-destructive, thermodynamic method for measuring the true molecular weight of proteins without external calibration. At equilibrium, the sedimentation and diffusion forces balance each other, leading to a concentration distribution that follows the equation:
Figure 2
M: Protein molecular weight
vˉ: Molar volume of the protein
ρ: Solution density
ω: Angular velocity
r: Centrifugal radius
T: Temperature
R: Gas constant
(1) Data Analysis
①The protein molecular weight is derived from the concentration gradient at equilibrium using nonlinear fitting methods.
②SE distinguishes different oligomeric states (e.g., monomer, dimer, trimer), though it is not suitable for highly aggregated samples.
(2) Applications
①High-precision molecular weight measurement: More accurate than SV, suitable for precise protein size analysis.
②Protein complex analysis: Effective for studying weakly interacting protein complexes, such as DNA-binding proteins and antibody-antigen complexes.
Ultracentrifugation Experimental Strategy and Key Technical Parameters
1. Sample Preparation
(1) Buffer Selection: High salt concentrations should be avoided, as they may affect the sedimentation behavior of proteins.
(2) Sample Purity: Proteins should be highly purified to avoid interference from impurities, which can impact the accuracy of the analysis.
(3) Concentration Range: Typically, concentrations range from 0.1 to 1 mg/mL; excessively high concentrations can reduce measurement accuracy.
2. Operating Parameters
(1) Centrifugal Speed: For Sedimentation Velocity (SV) experiments, speeds generally range from 40,000 to 60,000 rpm, while Sedimentation Equilibrium (SE) typically runs at 10,000 to 20,000 rpm.
(2) Temperature Control: Experiments are typically conducted at temperatures of 4°C or 20°C to maintain protein stability.
(3) Detection Method: The most common detection methods are ultraviolet absorption (UV 280 nm) and interference optical detection, both of which are used to record the data. UV 280 nm absorption is widely utilized for protein analysis, as it quantifies proteins based on the absorbance by aromatic amino acids.
Application of Ultracentrifugation in Protein Molecular Weight Determination
1. Studying the Native Conformation of Proteins
Sedimentation Equilibrium (SE) can be used to determine whether a protein maintains its native monomeric state in solution or forms dimers, tetramers, or higher-order oligomers. For example, certain proteins (such as P53) may oligomerize under different conditions, and ultracentrifugation can detect this phenomenon.
2. Analyzing Protein Complexes
Ultracentrifugation is utilized to study protein-protein or protein-small molecule interactions, such as antigen-antibody complexes, protease-inhibitor interactions, and more.
3. Determining the True Molecular Weight of Glycoproteins
Glycosylation modifications in glycoproteins can affect molecular weight measurements obtained via SDS-PAGE. SE provides the true molecular weight of the unglycosylated portion of the protein, offering a more accurate result.
4. Studying Protein Stability
By monitoring changes in the sedimentation coefficient, ultracentrifugation can assess whether the protein undergoes denaturation, dissociation, or aggregation.
Advantages and Limitations of Ultracentrifugation
1. Advantages
(1) Label-free, Non-denaturing: Ultracentrifugation requires no dyes or denaturing reagents, enabling the direct measurement of proteins in their native state.
(2) High Precision: SE offers highly accurate molecular weight measurements that are close to the true value.
(3) Studying Protein Interactions: It allows for the analysis of protein-protein and protein-nucleic acid complex stability.
2. Limitations
(1) Expensive Equipment: Ultracentrifuges are costly, and the technique requires professional training to operate.
(2) Long Experimental Duration: SE experiments typically require between 12 to 48 hours to complete.
(3) Complex Data Analysis: The data generated must be analyzed using specialized software, such as SEDFIT or SEDFIT-MSTAR, to perform fitting calculations.
Ultracentrifugation is a high-precision, non-destructive method for protein molecular weight determination, widely used in protein structure research, protein-ligand interaction analysis, and drug development. With advances in computational fitting technologies and the development of new ultracentrifugation instruments, this technique is poised to play an increasingly important role in protein research. Furthermore, combining ultracentrifugation with techniques such as Multi-Angle Light Scattering (MALS) and Isothermal Titration Calorimetry (ITC) will further enhance the accuracy and applicability of protein molecular weight determinations. At MtoZ Biolabs, we provide efficient ultracentrifugation for protein molecular weight determination services, leveraging cutting-edge technology platforms. We are committed to helping our clients uncover the biological functions of proteins through precise data analysis. Researchers with related needs are welcome to contact us for collaboration and innovation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?