Tunable Resistive Pulse Sensing (TRPS) based Exosome Characterization Service
- Dynamic Light Scattering (DLS) provides ensemble-averaged measurements, making it unable to distinguish apoptotic bodies and protein aggregates.
- Nanoparticle Tracking Analysis (NTA) offers size distribution but suffers from >30% concentration measurement error and lacks surface charge characterization.
- Size Distribution: High-resolution analysis of individual exosome diameters to distinguish subpopulation heterogeneity.
- Particle Concentration: Direct quantification of exosome numbers per unit volume, eliminating the need for standard curves.
- Surface Charge (Zeta Potential): Evaluation of membrane stability and intercellular interaction potential via electrophoretic mobility analysis.
Exosomes, 30-150 nm extracellular vesicles, transport proteins, nucleic acids, and lipids, playing crucial roles in tumor metastasis, immune regulation, and neurodegenerative diseases. With the rapid advancements in exosome-based biomarker discovery and drug delivery systems, there is an increasing demand for precise characterization of nanoscale exosome subpopulations.
Traditional methods face significant limitations:
These technological bottlenecks have hindered exosome research, leaving it in a “black box” stage, where quantitative models linking size, charge, and function remain undeveloped. Tunable Resistive Pulse Sensing (TRPS) has emerged as the new gold standard in exosome characterization, offering single-particle resolution and multiparametric analysis.
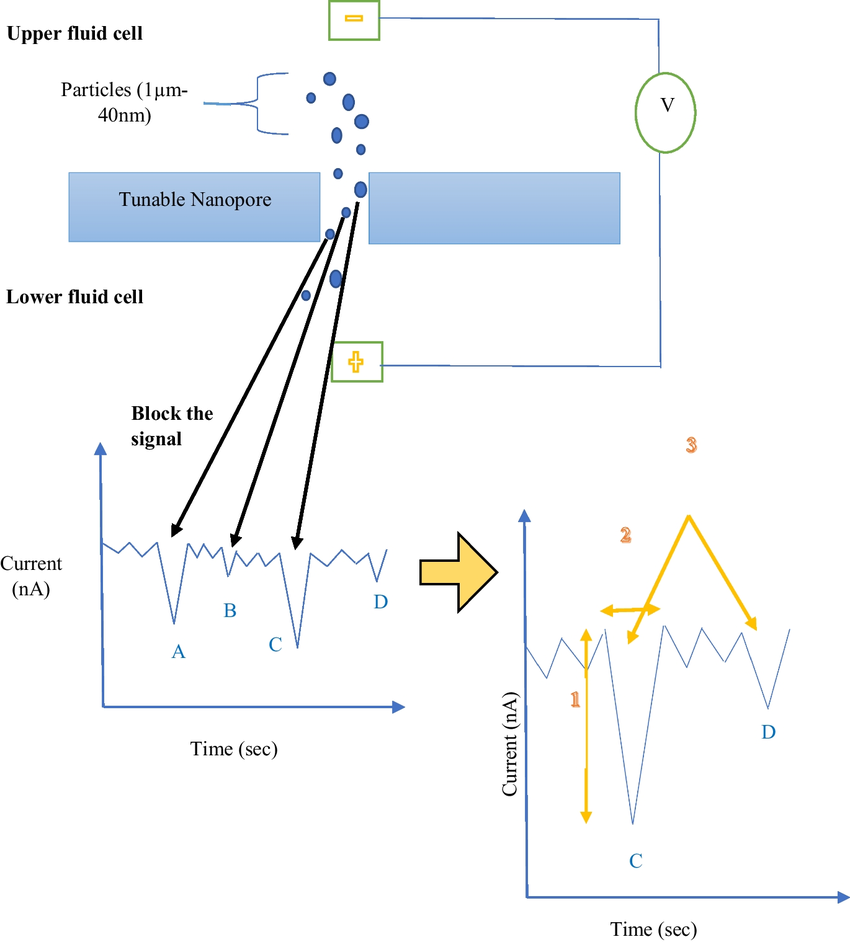
Chaturvedi, et al. Journal of Nanoparticle Research. 2024.
Figure1. Schematic representation of principle and working of tunable resistive pulse sensing (TRPS)
MtoZ Biolabs' TRPS-Based Exosome Characterization Service
In exosome research, TRPS technology has become an essential tool for high-resolution nanoparticle size and concentration analysis. However, its potential is often constrained by challenges such as pore clogging, background noise interference, and low sensitivity in complex biological samples.
Leveraging years of multi-omics exosome research experience, MtoZ Biolabs has optimized TRPS systems for stability and precision. We have significantly improved detection stability and signal-to-noise ratio, ensuring accurate subpopulation detection of 30-2000 nm exosomes across complex biological samples, including serum and cerebrospinal fluid. MtoZ Biolabs Tunable Resistive Pulse Sensing (TRPS)-based Exosome Characterization Service offers single-particle resolution and multiparametric detection, providing high-precision, reproducible, and standardized solutions for biopharmaceutical companies, research institutions, and clinical diagnostic centers.
Key Analytical Parameters
By integrating cutting-edge TRPS technology with standardized analytical workflows, MtoZ Biolabs provides researchers with unparalleled accuracy and insight into exosome biology, facilitating breakthroughs in biomarker discovery, drug delivery, and disease diagnostics.
Technical Principle
TRPS enables precise quantification of exosome physical characteristics through finely engineered nanopores. The core principle is as follows:
When exosomes pass through a nanopore with adjustable aperture under an electric field, they momentarily block the ionic current in the electrolyte solution, generating a resistive pulse signal that is proportional to the particle volume. By dynamically adjusting the pore size (50-2000 nm) and fluid pressure, TRPS ensures that exosomes of different sizes pass through the detection zone one by one, preventing signal overlap. At the same time, by analyzing the migration rate of particles in the electric field, TRPS can also calculate the surface Zeta potential, which provides critical parameters for evaluating exosome membrane stability and targeted delivery efficiency.
Compared to traditional techniques, TRPS' intelligent pore matching mechanism and single-particle counting capability allow it to maintain <5% concentration detection error even in complex biological samples (e.g., serum and cerebrospinal fluid).
Applications
1️⃣ Exosome-Based Drug Development: Monitor size uniformity of drug-loaded exosomes to ensure batch-to-batch consistency.
2️⃣ Disease Biomarker Discovery: Detect proportional shifts in exosome subpopulations under pathological conditions.
3️⃣ Engineered Exosome Optimization: Evaluate how surface modifications (e.g., antibody conjugation) influence zeta potential and stability.
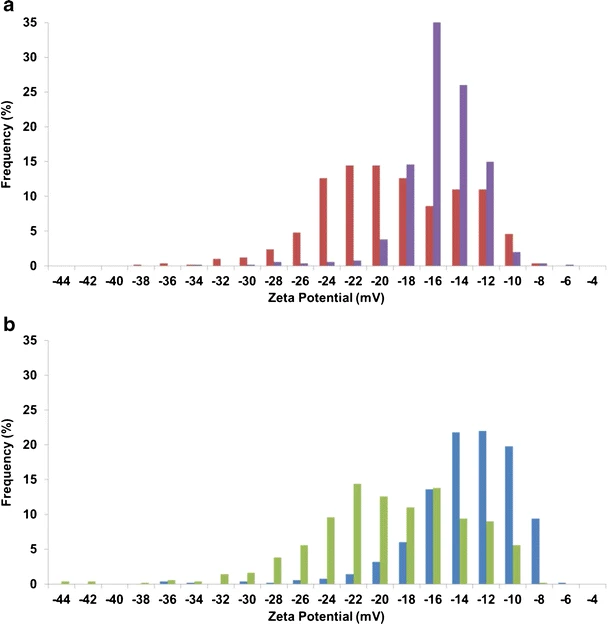
Blundell. et al. Anal Bioanal Chem. 2016.
Figure2. Characterizing the Changes and Distribution of Surface Charge on Protein Corona Using Tunable Resistive Pulse Sensing
Why Choose MtoZ Biolabs?
As a leader in multi-omics exosome solutions, MtoZ Biolabs’ TRPS-Based Exosome Characterization Service integrates TRPS, single-cell sequencing, and spatial proteomics to deliver:
✔ End-to-End Service: From exosome isolation and purification to multiparametric characterization and functional validation.
✔ Industrial-Grade Database: A size-charge correlation model covering 15 cancer types and 2000+ samples, accelerating biomarker discovery.
✔ Standardized Reporting: Provides GLP-compliant reports with size distribution histograms, concentration statistics, and charge analysis.
✔ Broad Dynamic Range: Compatible with complex biological samples (e.g., serum, cell supernatants), reducing purification requirements.
✔ Multiparametric Analysis: Simultaneous acquisition of size distribution, absolute concentration, and zeta potential in a single experiment.
✔ Full Size Spectrum Coverage: Accurately distinguishes 30 nm exosome subpopulations up to 2000 nm apoptotic bodies.
FAQs
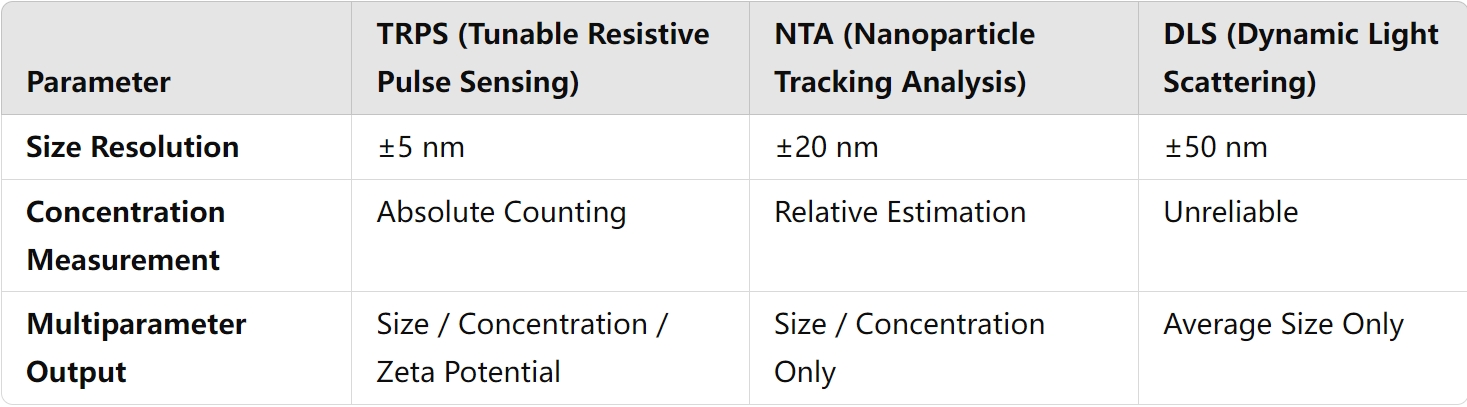
🔹 Q1: What are the core differences between TRPS and NTA?
A: TRPS achieves single-particle absolute counting via resistive pulse sensing, whereas NTA relies on light scattering for relative estimation. In the 10⁷-10¹⁰ particles/mL concentration range, TRPS has a detection error of <5%, significantly outperforming NTA (typical error 20-50%).
🔹 Q2: How much sample is required for analysis?
A: Minimum requirement: 200 μL (concentration ≥ 1×10⁸ particles/mL). For rare samples (e.g., cerebrospinal fluid), we offer an ultra-low volume enrichment protocol capable of processing as little as 50 μL.
How to order?







