The Hot Topic in Chromatin Biology: Insights into Histone Modification and CUT&Tag
Histone modifications are essential regulators of chromatin structure and gene expression, playing crucial roles in maintaining cellular function, genomic stability, and facilitating processes such as cell differentiation. Within the cell nucleus, DNA directs the synthesis of RNA and proteins, ensuring proper organismal function. DNA exists as a double helix and wraps around histones to form nucleosomes, which serve as the fundamental units of chromatin. Nucleosomes are organized into a bead-on-a-string structure and are further compacted to form chromatin, which is subsequently condensed into chromosomes during cell division. Throughout this process, histone modifications (such as acetylation, methylation, and phosphorylation) regulate chromatin accessibility, ultimately modulating gene expression. CUT&Tag (Cleavage Under Targets and Tagmentation) is a state-of-the-art technology that employs specific antibodies to target proteins or histone modification markers. Using Tn5 transposase (Tagmentase), this technique precisely fragments DNA at the target regions and simultaneously integrates sequencing adapters, enabling high-resolution mapping of transcription factor binding sites and histone modification landscapes across the genome.
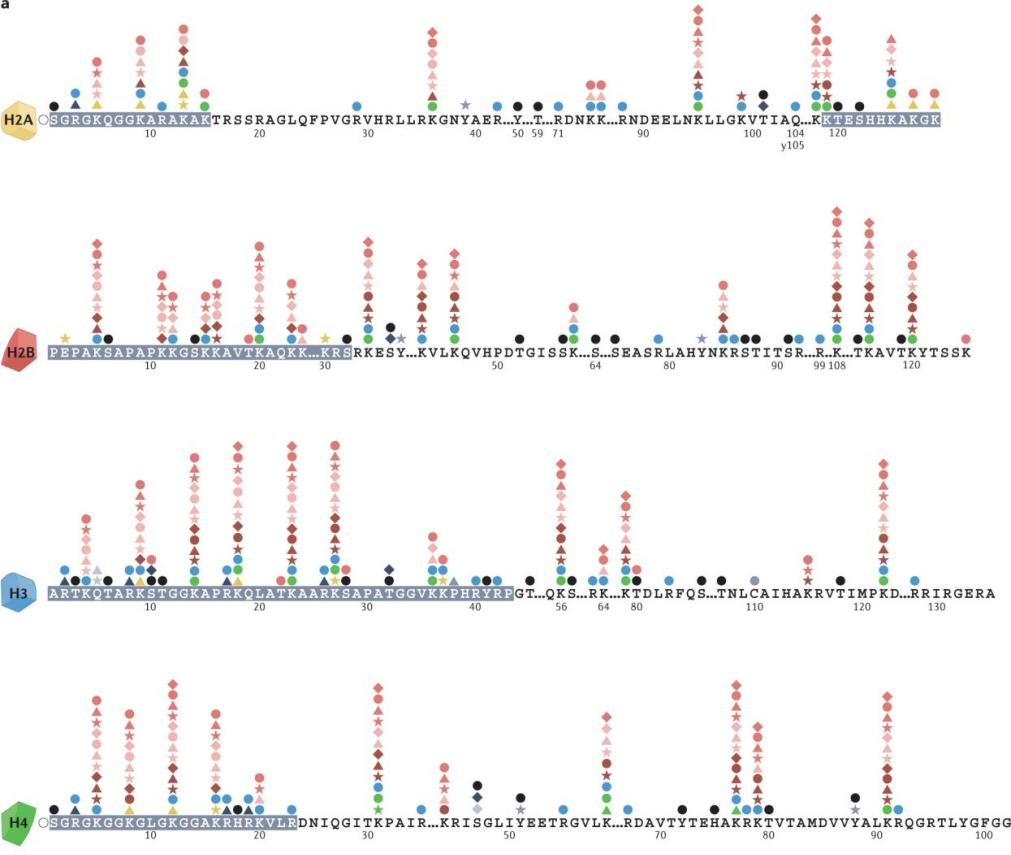
Figure 1. Different Histone Post-Translational Modification Sites (Nat Rev Genet 23, 563–580 (2022))
Traditional approaches such as ChIP-seq (Chromatin Immunoprecipitation Sequencing) have been instrumental in studying protein-DNA interactions and histone modification mechanisms. However, these methods suffer from significant limitations, including high sample input requirements, low accuracy, poor reproducibility, and suboptimal signal-to-noise ratios. In 2019, the Henikoff laboratory developed CUT&Tag (Nature Communications), a technology that significantly reduces sample input while improving signal quality and detection reliability. CUT&Tag leverages specific antibody recognition and high-throughput sequencing to generate precise genome-wide histone modification maps, offering insights into chromatin remodeling and transcription regulation.
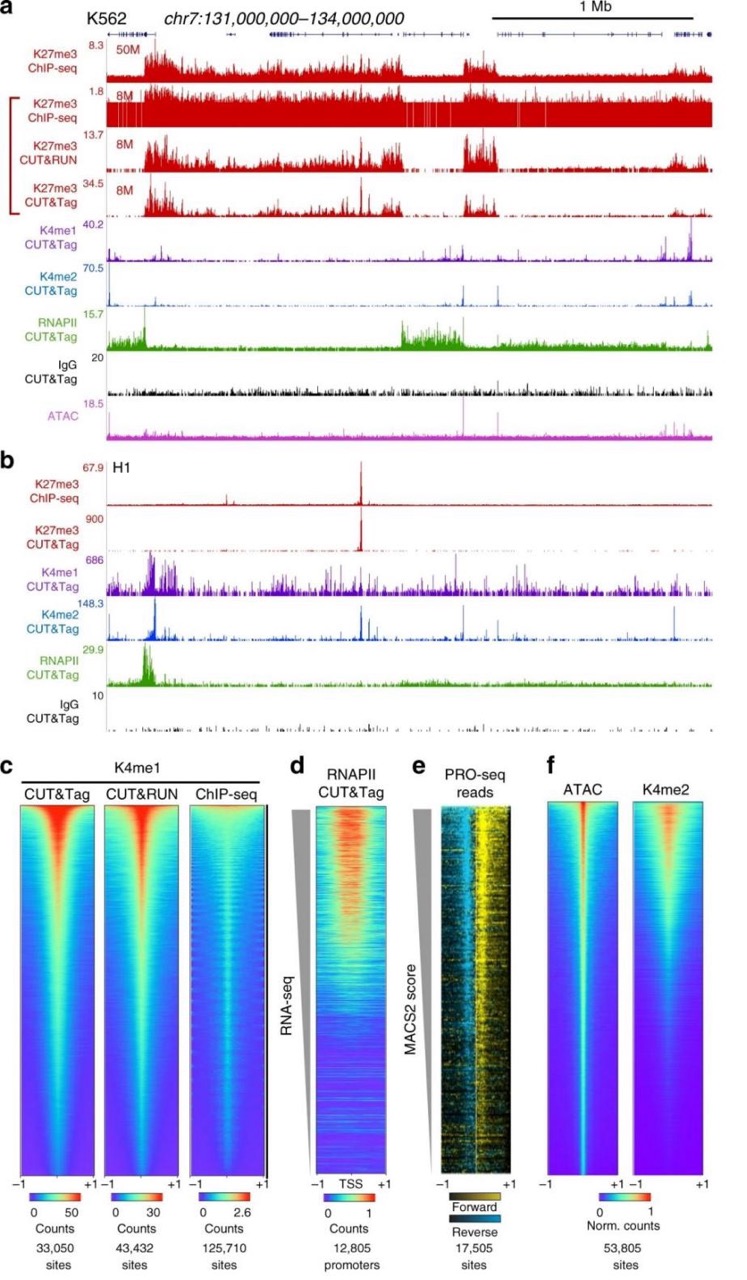
Figure 2. Comparison Between CUT&Tag and ChIP-seq Data (Nat Commun 10, 1930 (2019))
Analysis Workflow of CUT&Tag
1. Nucleus Extraction and Bead Binding
Both live cells and isolated nuclei can be used. ConA magnetic beads bind to glycoproteins on the cell membrane, and digitonin permeabilizes the membrane to facilitate downstream processes.
2. Primary and Secondary Antibody Binding
A primary antibody targets the protein of interest. Following incubation, cells are washed with a digitonin-containing buffer and incubated with a secondary antibody.
3. ChiTag Transposome Binding
The Protein A/G-Tn5 complex facilitates the binding of Tn5 transposase to the antibody-protein complex.
4. Tn5 Activation and DNA Fragmentation
In the presence of a reaction buffer containing Mg²⁺, Tn5 transposase becomes activated, fragmenting DNA precisely at the protein-binding sites while simultaneously attaching sequencing adapters to the fragmented DNA.
5. Sequencing and Data Analysis
The workflow includes data quality control, reference genome alignment, fragment analysis, peak enrichment analysis, motif discovery, and comparative peak enrichment analysis.
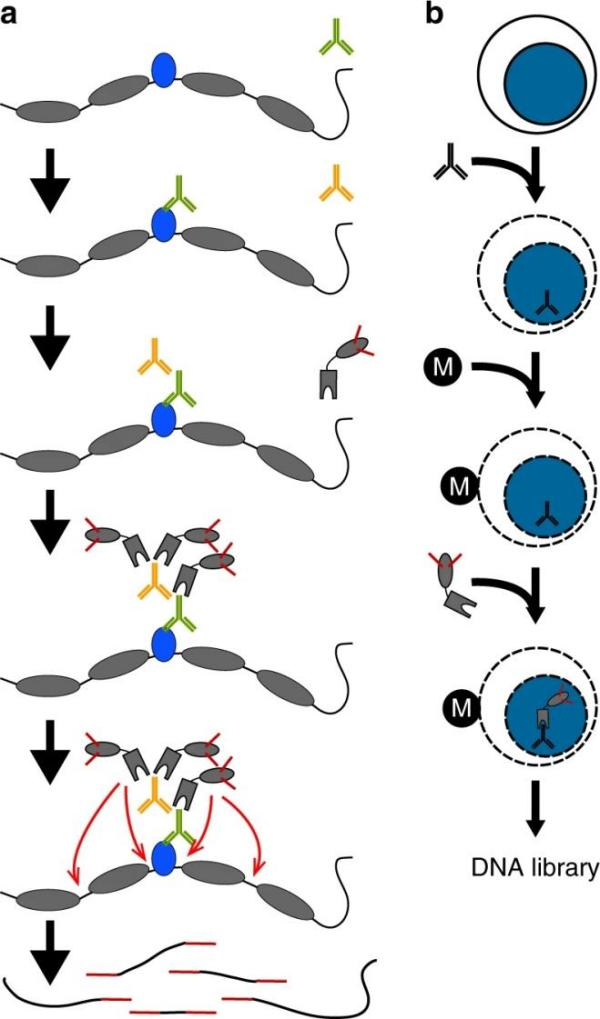
Figure 3. Analysis Workflow of CUT&Tag (Nat Commun 10, 1930 (2019))
Application Reference 1
Title: Positive Feedback Regulation of Microglial Glucose Metabolism by Histone H4 Lysine 12 Lactylation in Alzheimer’s Disease
Journal: Cell Metabolism
Impact Factor: 27.7
Year: 2022
Research Summary:
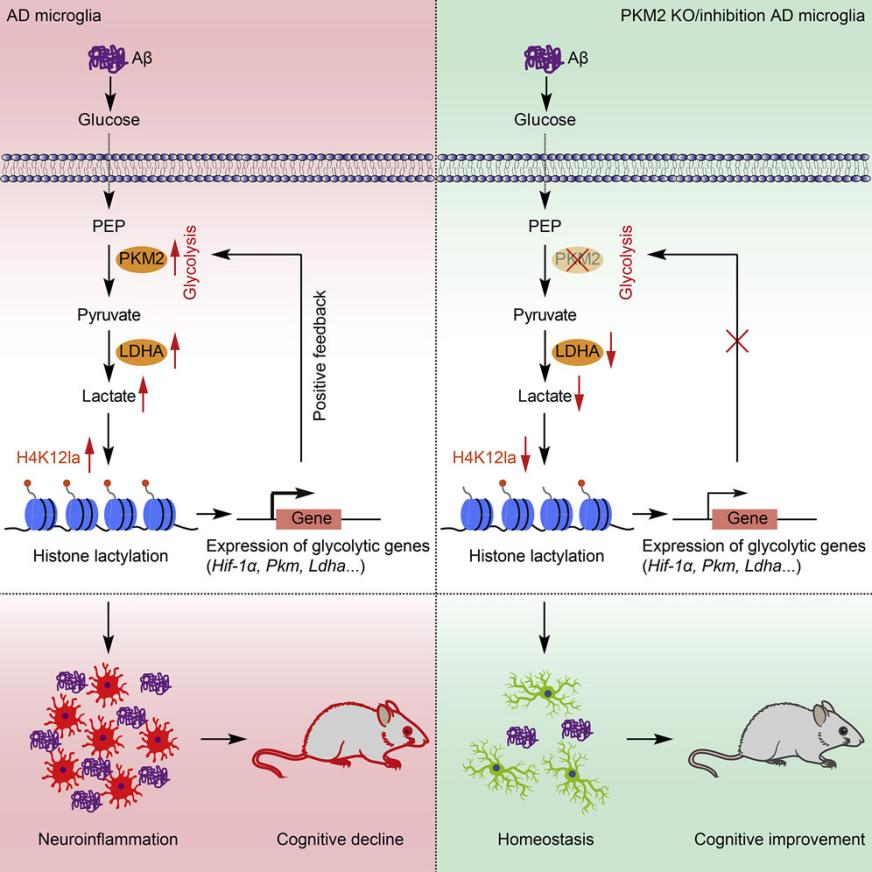
Figure 4.
Research Content: Pro-inflammatory activation of microglia is a hallmark of Alzheimer’s disease (AD), characterized by a metabolic shift from oxidative phosphorylation (OXPHOS) to glycolysis. This study identifies a glycolysis-driven positive feedback loop in microglia that exacerbates AD pathology. Elevated levels of histone lactylation at H4K12 (H4K12la) were observed in both 5XFAD mouse models and human AD brain samples, specifically in microglia adjacent to Aβ plaques. This lactate-dependent histone modification accumulates on the promoters of glycolytic genes, promoting their transcription and enhancing glycolytic activity. The resulting glycolysis/H4K12la/PKM2 positive feedback loop contributes to microglial dysfunction and worsens AD pathology. Importantly, pharmacological inhibition of PKM2 or microglia-specific deletion of PKM2 reduced microglial activation, alleviated Aβ burden, and improved cognitive function in AD mouse models. These findings suggest that targeting this feedback loop could serve as a potential therapeutic strategy for treating AD.
Application Reference 2
Title: A Feedback Loop Driven by H3K9 Lactylation and HDAC2 in Endothelial Cells Regulates VEGF-Induced Angiogenesis
Journal: Genome Biology
Impact Factor: 10.1
Year: 2024
Research Summary:
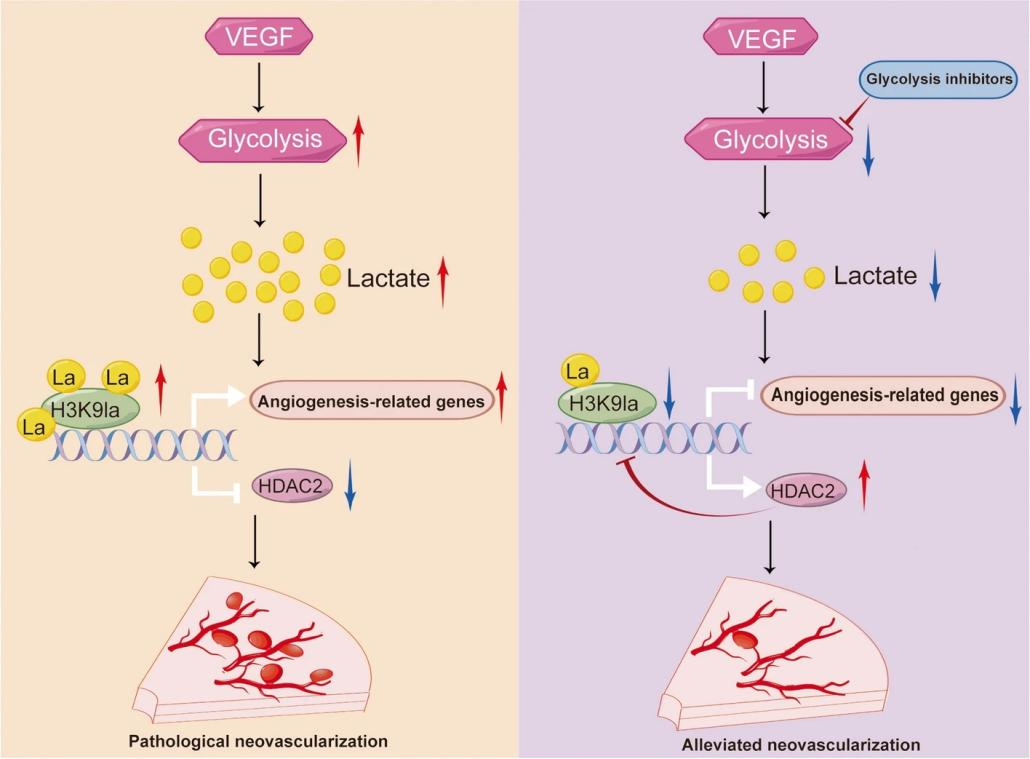
Figure 5.
Research Content: Vascular endothelial growth factor (VEGF) is a critical pro-angiogenic factor involved in various pathological conditions. Enhanced glycolysis and lactate accumulation are strongly associated with pathological angiogenesis. This study identified a feedback loop between H3K9 lactylation (H3K9la) and histone deacetylase 2 (HDAC2) that drives VEGF-induced angiogenesis. VEGF stimulation elevated H3K9la levels in endothelial cells, while pharmacological inhibition of glycolysis suppressed H3K9la and impaired angiogenesis. CUT&Tag analysis demonstrated H3K9la enrichment on angiogenesis-related gene promoters, enhancing their transcription. Notably, increased H3K9 acetylation suppressed HDAC2 expression, while HDAC2 overexpression reduced H3K9 acetylation and inhibited angiogenesis. These findings suggest that targeting the H3K9la/HDAC2 feedback loop could be a promising therapeutic strategy for pathological angiogenesis.
Continual advancements in histone modification and CUT&Tag analysis have enabled researchers to uncover the intricate relationships between chromatin structure dynamics and gene expression regulation with remarkable precision. These insights form a robust molecular foundation for studying disease mechanisms, drug discovery, and personalized therapies. MtoZ Biolabs offers histone modification and CUT&Tag analysis services with high sensitivity, reproducibility, and low-input requirements, supported by a highly experienced technical team and advanced quality control platforms. These services enable researchers to generate high-resolution genome-wide histone modification maps, advancing their understanding of gene regulatory networks and supporting efficient project outcomes.
For further details about MtoZ Biolabs' histone modification and CUT&Tag analysis services, please contact our technical support team for professional assistance.
References
[1] Millán-Zambrano G, Burton A, Bannister AJ, Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet. 2022 Sep;23(9):563-580. doi: 10.1038/s41576-022-00468-7. Epub 2022 Mar 25. PMID: 35338361.
[2] Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019 Apr 29;10(1):1930. doi: 10.1038/s41467-019-09982-5. PMID: 31036827; PMCID: PMC6488672.
[3] Pan RY, He L, Zhang J, Liu X, Liao Y, Gao J, Liao Y, Yan Y, Li Q, Zhou X, Cheng J, Xing Q, Guan F, Zhang J, Sun L, Yuan Z. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer's disease. Cell Metab. 2022 Apr 5;34(4):634-648.e6. doi: 10.1016/j.cmet.2022.02.013. Epub 2022 Mar 17. PMID: 35303422.
[4] Fan W, Zeng S, Wang X, Wang G, Liao D, Li R, He S, Li W, Huang J, Li X, Liu J, Li N, Hou S. A feedback loop driven by H3K9 lactylation and HDAC2 in endothelial cells regulates VEGF-induced angiogenesis. Genome Biol. 2024 Jun 25;25(1):165. doi: 10.1186/s13059-024-03308-5. PMID: 38918851; PMCID: PMC11197246.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







