Target Identification and Validation Service| Drug Discovery
-
CAR-T Target Expansion
Overcoming reliance on CD19 and enabling discovery of new targets for solid tumors. -
Monoclonal Antibody Development
Identifying novel antigens with high specificity and affinity for antibody therapeutics. -
Personalized Oncology
Providing individualized target profiles to support precision cancer therapy. -
Immune Checkpoint Inhibitor Discovery
Identifying new membrane-bound immune modulators for next-generation checkpoint therapies. -
Drug Repurposing
Revealing new therapeutic targets for existing compounds, enabling repositioning strategies.
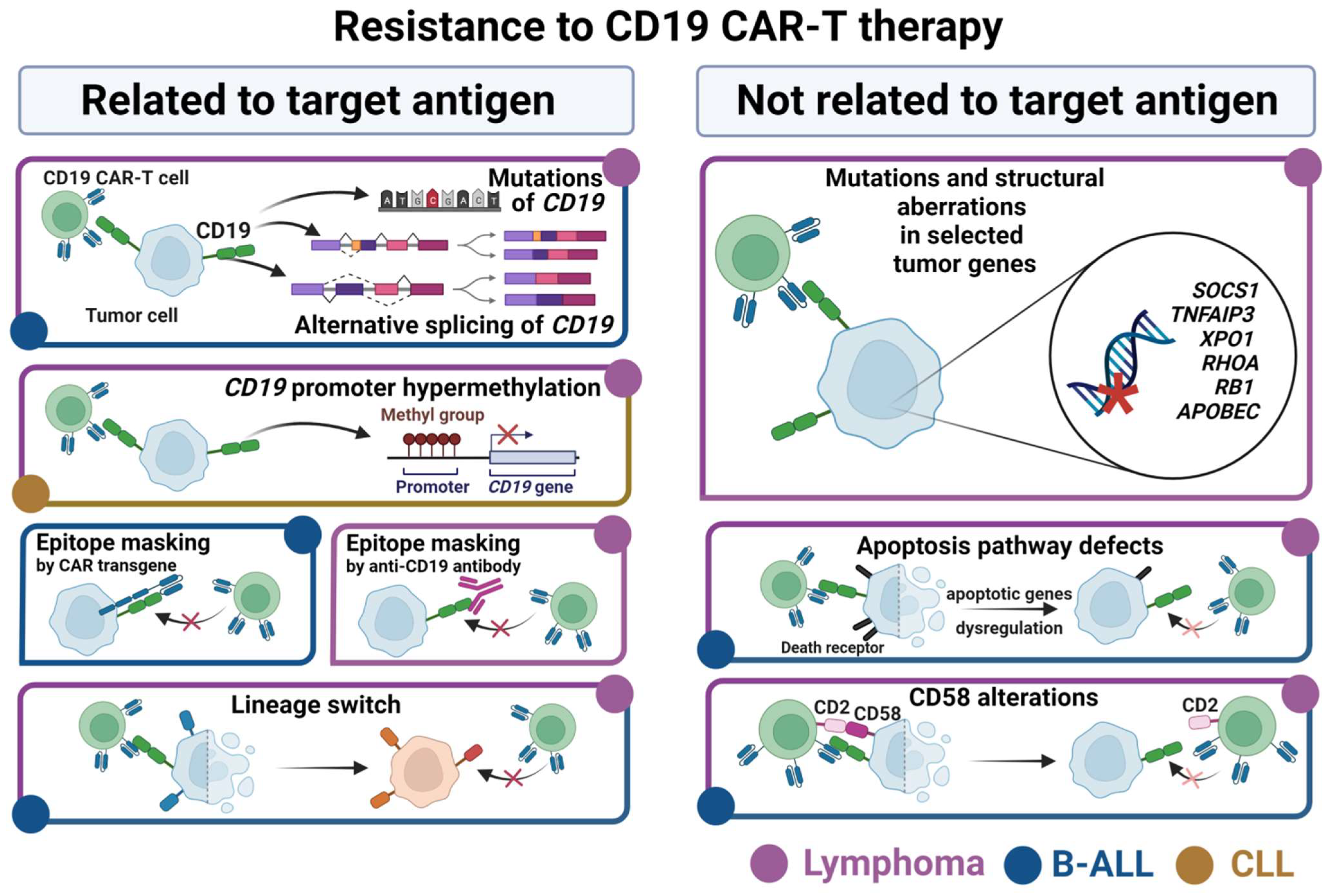
The advent of cancer immunotherapy has revolutionized oncology, with chimeric antigen receptor T-cell (CAR-T) therapy standing out as a particularly transformative modality. CAR-T therapy involves genetically engineering autologous T cells to express synthetic receptors that can specifically recognize tumor-associated antigens, enabling precise and potent elimination of malignant cells. However, current clinical trials and applications are overwhelmingly centered on a limited set of antigens—most notably CD19, which is the target in over 40% of CAR-T trials globally. This over-reliance on a narrow antigen spectrum has resulted in a shortage of new, effective, and specific targets, thereby limiting the broader advancement of CAR-T therapies and the immunotherapeutic drug industry.
The discovery of novel tumor-specific targets is critical not only for overcoming the limitations of target redundancy but also for guiding innovative therapeutic design and expanding treatment options. The Human Membrane Protein Array (HMPA) represents a cutting-edge, high-throughput, and highly sensitive platform capable of rapidly identifying druggable surface antigens from primary tumor cells, making it a key technology to address this pressing bottleneck in immunotherapy development.
Marhelava, K. et al. Cells. 2022.
Figure1. B Cell-Derived Cancers Develop Various Mechanisms of Resistance to CD19 CAR-T Therapy
Service at MtoZ Biolabs
Leveraging the Human Membrane Protein Array (HMPA) platform in combination with ultra-sensitive mass spectrometry, MtoZ Biolabs offers a comprehensive target identification and validation service—covering target screening, specificity analysis, and functional validation. Supported by extensive membrane protein chip libraries, a dedicated technical team, and robust bioinformatics pipelines, we empower researchers to efficiently identify and validate therapeutic targets. Whether you are developing monoclonal antibodies, CAR-X cell therapies, or next-generation immunotherapies, MtoZ Biolabs provides the critical technical infrastructure to accelerate your discovery pipeline and drive therapeutic innovation forward.
Technical Principles
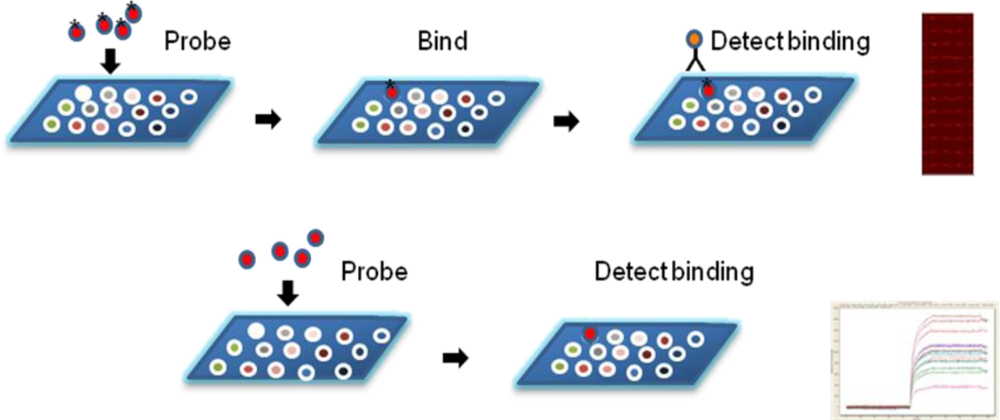
The HMPA is a high-content analytical technology designed to assess interactions between thousands of human plasma membrane proteins and candidate therapeutics (e.g., monoclonal antibodies, CAR-T receptors). By expressing full-length membrane proteins on specially designed carriers or microarrays, the system preserves native conformations and biological activity.
Candidate antibodies or CAR-T receptors are directly incubated with the immobilized membrane proteins. Specific interactions are detected using high-sensitivity fluorescence labeling and imaging, allowing real-time acquisition of binding signals. Signal intensity and localization analysis provide critical information on target specificity and affinity, enabling rapid screening and accurate identification of promising new targets for therapeutic development.
Natesan, M. et al. Int. J. Mol. Sci. 2010.
Figure2. Measuring Probe Interactions with Microarrayed Proteins
Analysis Workflow
MtoZ Biolabs implements a streamlined and standardized workflow to ensure accurate, reproducible, and biologically meaningful target discovery. The key steps include:
1. Primary Tumor Cell Preparation
Primary tumor cells are isolated and purified from patient tissue or clinical samples. This ensures the preservation of biological relevance and high cell viability.
2. Membrane Protein Array Construction
Human plasma membrane proteins are overexpressed using a stable expression system and immobilized on chip surfaces while maintaining structural integrity and bioactivity.
3. Candidate Drug Interaction Detection
Candidate agents (e.g., antibodies or CAR receptors) are incubated with the array. Interactions are detected using high-resolution fluorescence labeling and imaging, and both signal intensity and spatial binding data are recorded.
4. Data Analysis and Target Confirmation
Advanced bioinformatics tools are used to mine interaction datasets. Biological validation experiments are performed to confirm the therapeutic relevance and druggability of candidate targets.
5. Target Validation and Functional Assessment
Candidate targets are further evaluated through in vitro and in vivo functional assays, including mechanistic studies and efficacy modeling, to verify their potential for downstream drug development.
Why Choose MtoZ Biolabs?
✔️ Technological Leadership
Powered by internationally advanced HMPA and high-sensitivity MS platforms for efficient and accurate target identification.
✔️Proven Experience
Extensive track record in successful target discovery and validation with flexible, tailored solutions.
✔️High-Quality Data
High-resolution detection ensures reliable, reproducible, and quantitative results.
✔️Efficient Workflow
One-stop integrated service model accelerates the pace of drug development.
✔️One-Time Charge
Applications
The novel target discovery service by MtoZ Biolabs is broadly applicable to various therapeutic development programs:
Frequently Asked Questions (FAQs)
Q1: How does the Human Membrane Protein Array differ from conventional protein microarrays?
A1: Unlike traditional arrays, which may include soluble or non-native proteins, the HMPA focuses specifically on full-length membrane proteins anchored in formats that preserve native conformation and biological function. This allows for physiologically relevant interaction screening and higher translational potential for drug development.
Q2: Is tumor tissue the only valid sample type for this analysis?
A2: While primary tumor samples are the main focus, the platform is also adaptable to other disease models, such as autoimmune or neurodegenerative conditions, enabling identification of disease-specific membrane protein targets.
Q3: Will the final deliverables include bioinformatics analysis?
A3: Yes. Each project includes a comprehensive, traceable analytical report, detailing protein-drug interaction maps, criteria for target selection, binding intensity scores, expression background information, and recommended next-step validation strategies—supporting seamless downstream R&D.
How to order?







