Sulfation Proteomics Service
Protein sulfation is a critical post-translational modification (PTM) involving the covalent addition of a sulfate group to specific amino acid residues, primarily tyrosine (Tyr) and occasionally serine (Ser) or threonine (Thr). Catalyzed by enzymes known as tyrosylprotein sulfotransferases (TPSTs), this modification typically occurs in the trans-Golgi network and uses 3'-phosphoadenosine-5'-phosphosulfate (PAPS) as the universal sulfate donor. Although less frequently discussed than phosphorylation or glycosylation, sulfation is essential for numerous biological processes, including protein–protein interactions, hormone activity, immune response modulation, extracellular matrix organization, and viral entry. It plays a prominent role in chemokine receptor function, blood coagulation, neurotransmission, and cell adhesion.
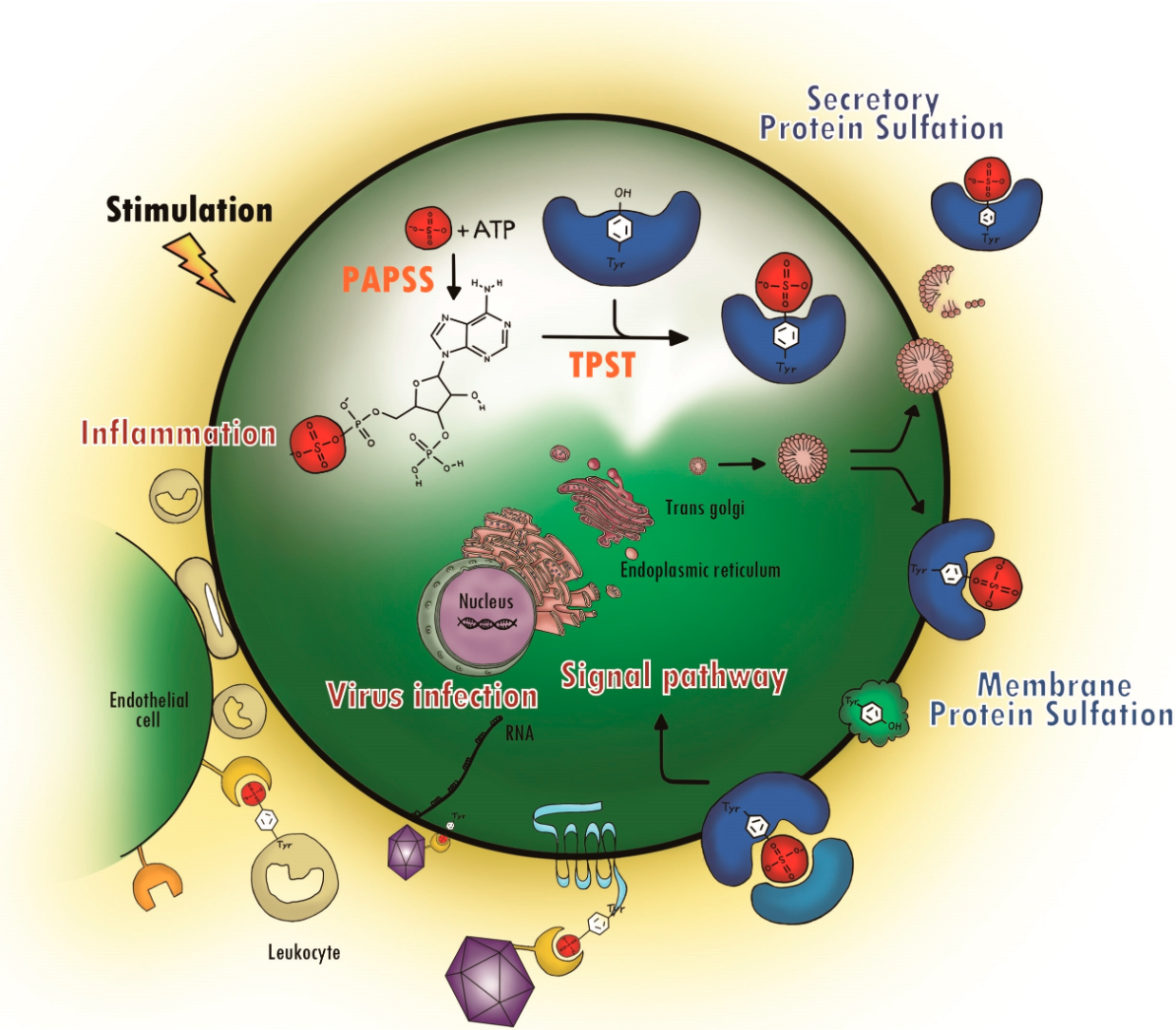
Figure 1. Protein Tyrosine Sulfation (PTS) and Its Biological Path
Recent studies have implicated aberrant sulfation in a range of pathological conditions, including cancer, inflammatory diseases, autoimmune disorders, and viral infections. However, sulfation remains one of the more technically challenging PTMs to detect and quantify due to the lability of sulfate groups and their low stoichiometry. To support researchers exploring this PTM, MtoZ Biolabs offers a specialized Sulfation Proteomics Service that combines optimized enrichment strategies, high-resolution LC-MS/MS, and expert data analysis to deliver accurate, site-specific, and quantitative sulfation profiles.
Analysis Workflow
With our advanced analytical platforms and specialized workflows, we enable precise identification, quantification, and functional annotation of sulfated proteins.
1. Sample Preparation
Proteins are extracted, denatured, reduced, alkylated, and digested with trypsin or other proteases. Peptide mixtures are desalted and quality checked.
2. Sulfated Peptide Enrichment
Enrichment is performed using chemical capture or antibody-based methods, depending on sample complexity and research goals.
3. LC-MS/MS Analysis
Enriched peptides are analyzed using nanoLC coupled to Orbitrap MS/MS platforms. ETD or EThcD fragmentation is used to preserve labile sulfate groups.
4. Data Processing
Raw data are analyzed with advanced software, using sulfate-specific variable modifications and accurate site localization scoring.
5. Functional Annotation
Identified sulfation sites are annotated for biological pathways, GO terms, protein–protein interactions, and PTM crosstalk.
Service Advantages
☑️Advanced Analysis Platform: MtoZ Biolabs established an advanced Sulfation Proteomics Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
☑️One-Stop Service: From sample preparation and sulfated peptide enrichment to LC-MS/MS analysis and bioinformatics interpretation, we deliver a fully integrated workflow that saves time and ensures data consistency.
☑️Customizable Experimental Design: We tailor every project based on your research goals, sample type, and target PTMs. Whether discovery-based profiling or targeted validation, our workflows are built to fit your needs.
☑️Experienced Scientific Team: Our proteomics specialists have extensive experience in post-translational modification analysis and provide expert guidance throughout your project—from experimental setup to data interpretation.
Applications
💠Immunology and Inflammation: Profile sulfated proteins involved in immune cell migration, antigen presentation, and cytokine signaling.
💠Infectious Disease Mechanisms: Investigate how sulfation facilitates pathogen binding and entry, particularly in viral infections where host receptors are modified.
💠Drug Development and Biotherapeutics: Characterize sulfation in therapeutic proteins, antibodies, or fusion constructs, ensuring batch consistency and biological activity.
💠PTM Crosstalk: Study the interplay between sulfation and other PTMs such as phosphorylation and glycosylation in complex signaling networks.
Case Study
Tyrosine Sulfation in Histone Regulation
Tyrosine sulfation is a common post-translational modification, but its role in histone regulation has been largely unexplored. In this study, SULT1B1 was reported as a histone sulfotransferase responsible for sulfating tyrosine 99 on histone H3 in the cytosol. This modification, H3Y99sulf, facilitates the transport of histone H3 into the nucleus, where it influences gene promoter regions and interacts with PRMT1 to promote H4R3me2a deposition, impacting gene transcription. Disrupting H3Y99sulf reduced PRMT1 binding and transcription levels. These findings highlight the importance of tyrosine sulfation in chromatin dynamics and transcriptional regulation, demonstrating how Sulfation Proteomics Service can provide essential insights into protein sulfation in complex biological systems.
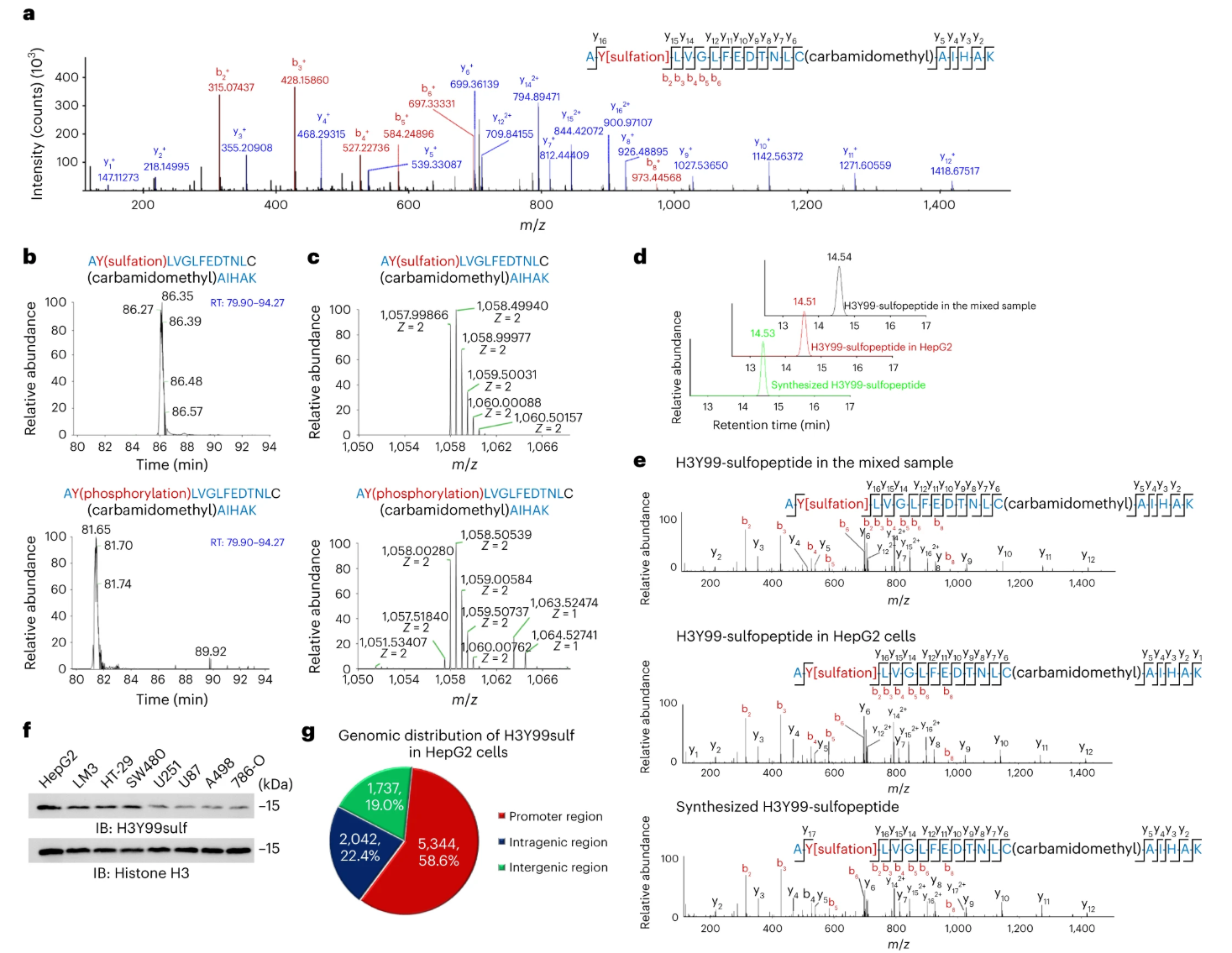
Figure 2. Comprehensive Analysis of Histone Tyrosine Sulfation (H3Y99sulf) in HepG2 Cells
FAQ
Q: What types of samples are suitable for sulfation analysis?
We accept a wide range of samples including cultured cells, tissues, plasma/serum, and purified proteins. All samples should be free of detergents and protease inhibitors to ensure compatibility with downstream enrichment and LC-MS/MS analysis.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Partner with MtoZ Biolabs for high-resolution, site-specific sulfation analysis tailored to your research goals. Contact us today to discuss your project or request a custom quote.
How to order?







