Single-Cell Transcriptomics Service
Single-cell transcriptomics is a cutting-edge technology that enables in-depth analysis of gene expression at the single-cell level, uncovering cellular heterogeneity and individual variability. Traditional transcriptomics techniques rely on bulk tissue data, which fail to capture the subtle differences between cells within the same tissue or sample. Single-cell transcriptomics overcomes this limitation, providing detailed gene expression profiles for individual cells and offering a more nuanced biological perspective.
Single-cell transcriptomics allows for the comprehensive analysis of various transcriptomic molecules, including mRNA and non-coding RNAs (e.g., lncRNA, miRNA, circRNA). It enables researchers to study dynamic cellular changes and functional differences under diverse biological conditions. With this technology, we can explore complex biological processes such as cell differentiation, development, and disease mechanisms, thus advancing fields like precision medicine and cancer research.
MtoZ Biolabs offers Comprehensive Single-Cell Transcriptomics Service supporting clients with everything from sample preparation to data analysis to obtain high-quality research outcomes. We utilize state-of-the-art platforms, including the 10x Genomics Chromium system combined with the Illumina platform, to ensure accurate and comprehensive solutions
Services at MtoZ Biolabs
At MtoZ Biolabs, we provide a range of Single-Cell Transcriptomics Service that allow you to investigate cellular functions and heterogeneity from various perspectives. These include, but are not limited to, the following services:
1. Single-Cell RNA Sequencing
Using high-throughput single-cell RNA sequencing technology, we analyze gene expression in individual cells, helping researchers identify distinct cell types and their roles in biological processes.
2. Single-Cell Non-coding RNA Sequencing
By identifying and analyzing the expression of various non-coding RNAs (e.g., lncRNA, miRNA, circRNA), we explore their roles in gene regulation and cellular function.
3. Spatial Transcriptomics
This technology integrates tissue sectioning with spatial information, allowing us to analyze gene expression differences within the cellular microenvironment and understand the relationships between cell distribution and function.
4. Single-Cell T Cell Receptor Sequencing
Single-cell T cell receptor (TCR) sequencing enables analysis of the diversity and immune response of T cells, providing valuable insights into T cell recognition of antigens, particularly in immunotherapy and autoimmune disease research.
5. Single-Cell Epigenomics
This service examines RNA modifications and their regulatory effects on gene expression and cellular function, providing insights into the dynamic changes of the epitranscriptome in disease contexts.
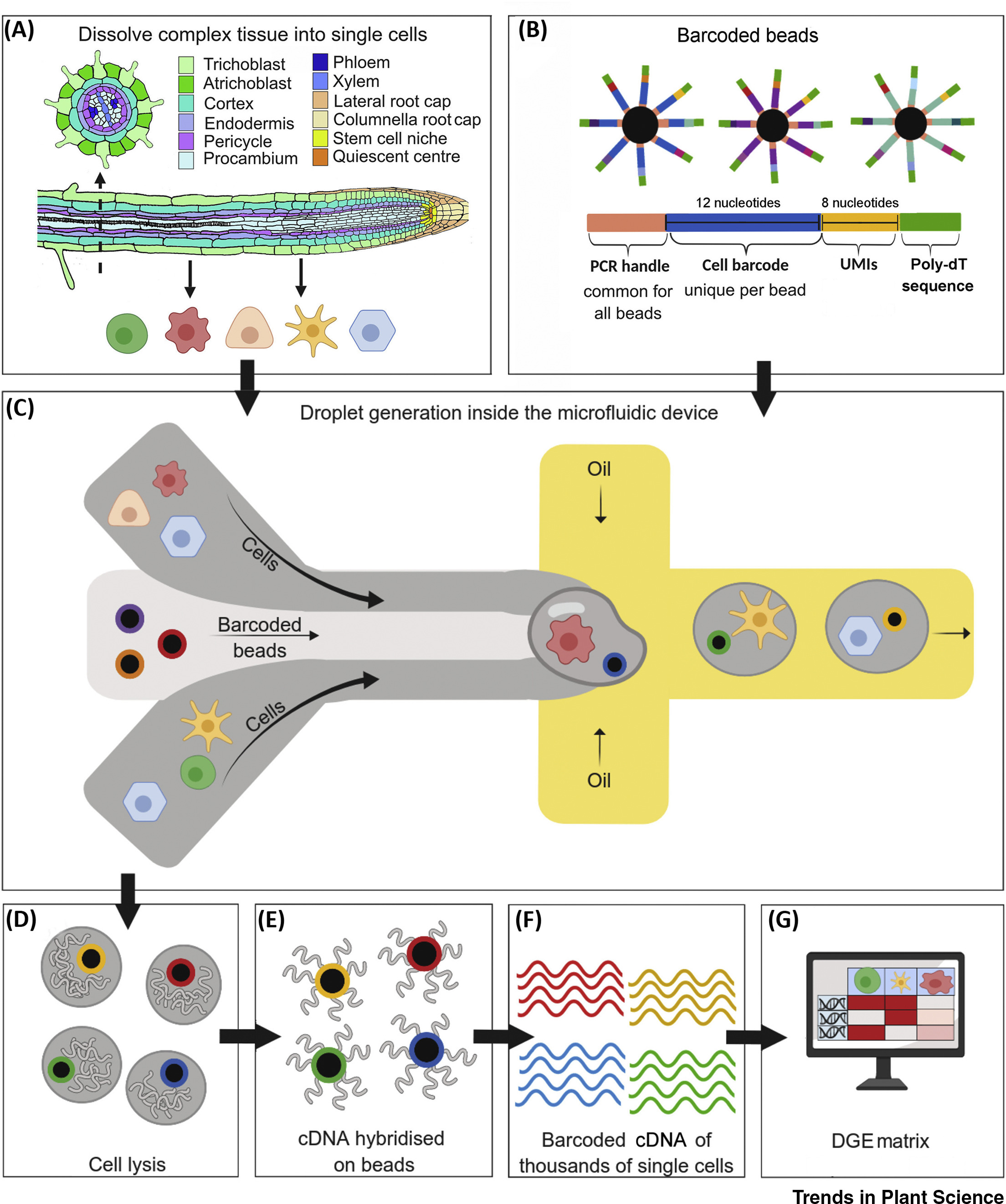
Rich-Griffin, C. et al. Trends Plant Sci. 2020.
Figure 1. The for Workflow for Singe-Cell Transcriptomics
Service Advantages
1. High Throughput with High Resolution
By combining the 10x Genomics platform with Illumina sequencing, we ensure high-throughput analysis while providing precise measurement of gene expression at the single-cell level.
2. Multi-dimensional Data Integration
Our Single-Cell Transcriptomics Service integrates mRNA, non-coding RNA, and epigenomics data, offering a comprehensive view of cellular transcriptomics and facilitating in-depth analysis of gene expression complexities.
3. Multi-Sample Compatibility
Our platform supports a wide variety of biological samples, including cells, tissues, blood, and bodily fluids, providing flexibility to accommodate diverse research demands.
4. One-Stop Solution
We provide full-service solutions from sample preparation to data analysis, streamlining research workflows and enhancing efficiency.
5. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
Case Study
Case1: Advancing Plant Functional Genomics with Single-Cell Transcriptomics
This study showcases how single-cell transcriptomics has revolutionized plant functional genomics by revealing plant cell type diversity, developmental dynamics, and gene regulatory networks. Single-cell transcriptomics's high-resolution capabilities have been instrumental in identifying cell-specific gene expression patterns, playing a significant role in studies of stress resistance and key gene function analysis.
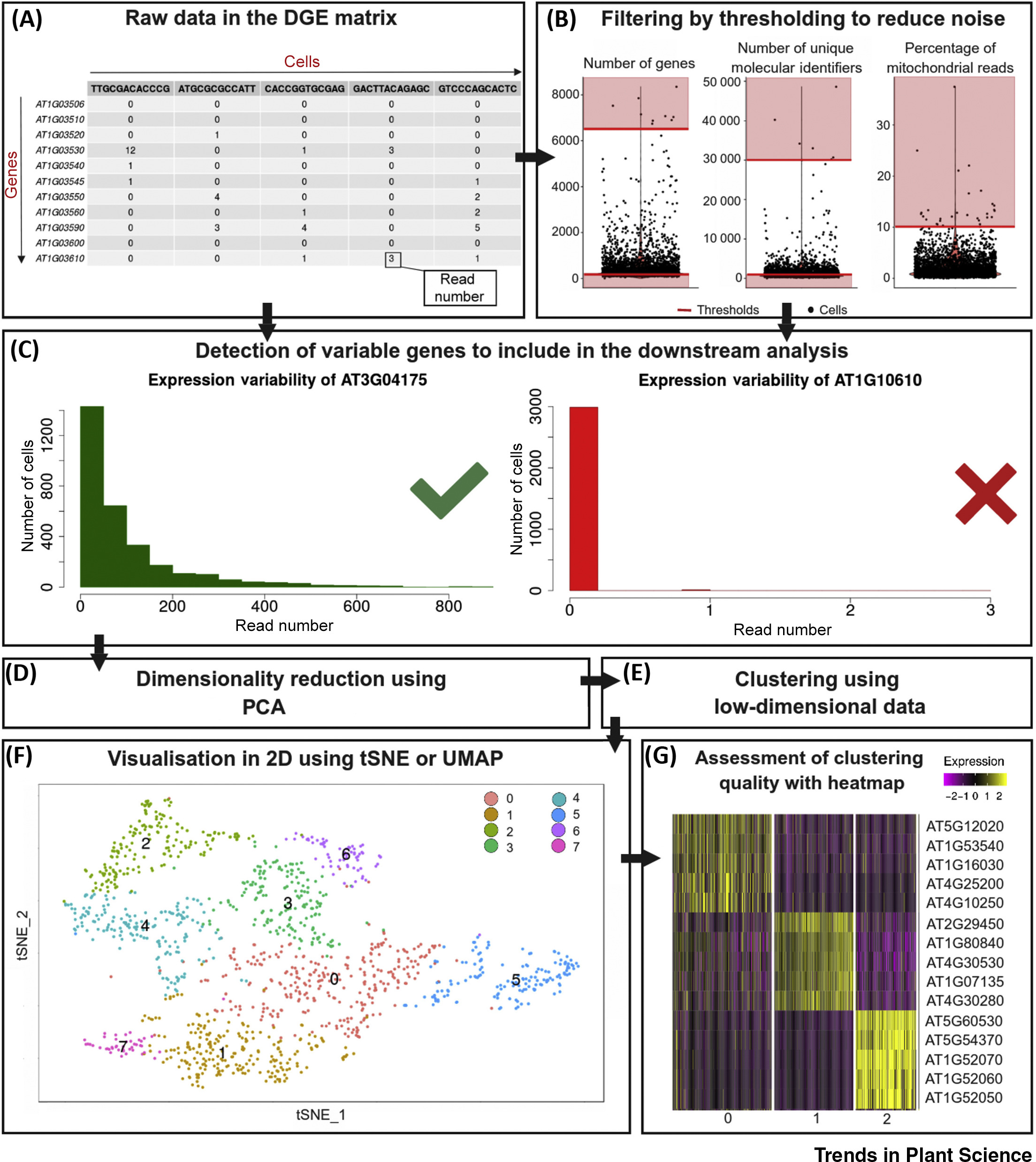
Rich-Griffin, C. et al. Trends Plant Sci. 2020.
Case 2: Single-Cell Transcriptomics in Mtb/HIV Co-Infection Research
This study explores the potential of single-cell transcriptomics in studying Mycobacterium tuberculosis (Mtb) and HIV co-infection. It highlights how this technology can track dynamic immune cell changes and identify key genes and subpopulations, underscoring its value in understanding immune suppression mechanisms and molecular pathology in infectious diseases.
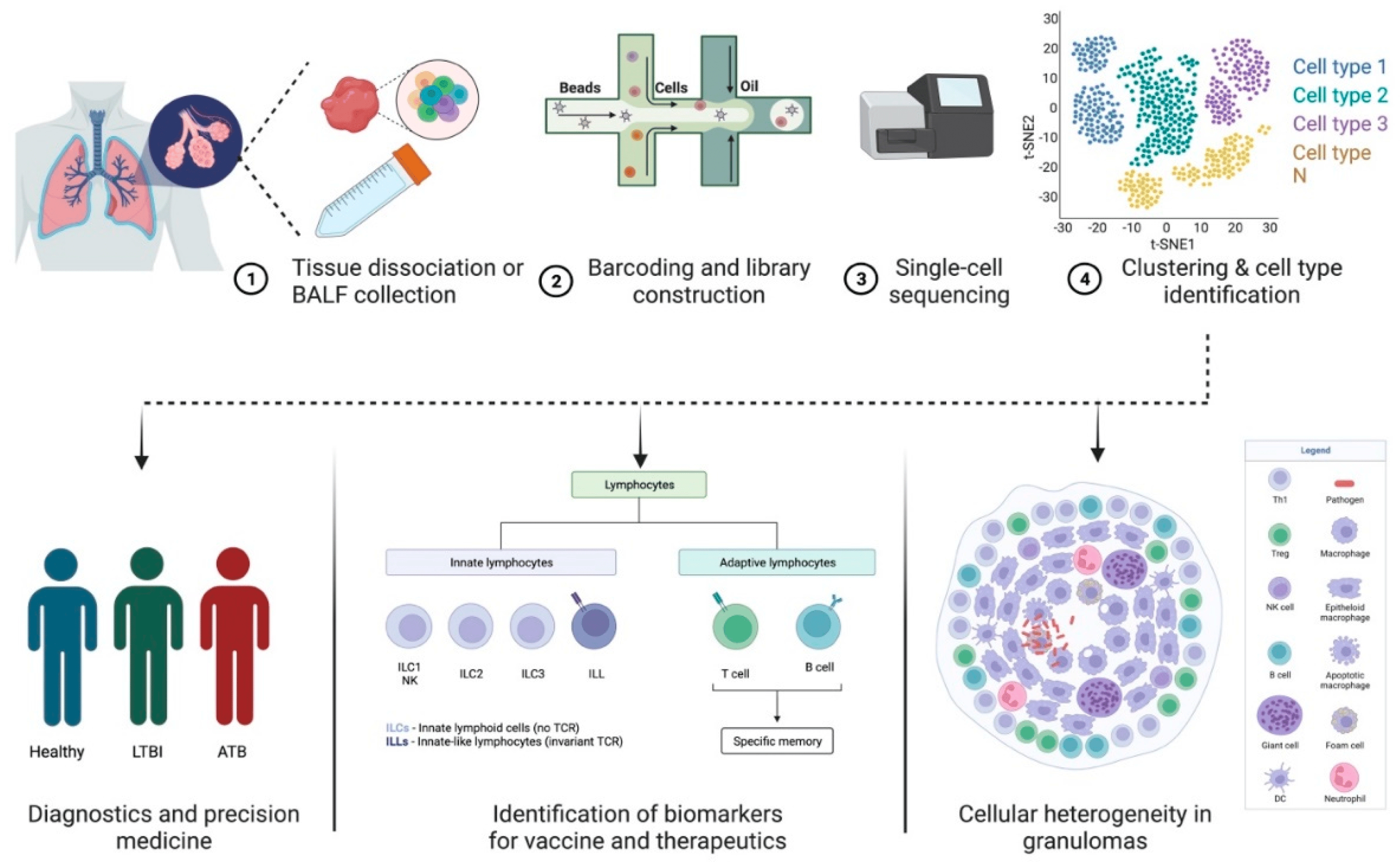
Kulkarni, S. et al. Cells. 2023.
Single-cell transcriptomics has broad applications across fields such as cell fate determination, developmental biology, immunology, oncology, and neuroscience. It enables deeper insights into cellular heterogeneity within the tumor microenvironment, immune responses, and key regulatory mechanisms in tissue development and regeneration.
MtoZ Biolabs' Single-Cell Transcriptomics Service utilizing advanced technologies and expert data analysis, provide a comprehensive, one-stop solution for researchers seeking precise insights in both cell biology and clinical studies.
For more details, feel free to contact us for a free consultation!
FAQ
Q1: What types of samples can I provide for single-cell transcriptomics analysis?
MtoZ Biolabs accepts a wide range of sample types, including raw tissue, cell suspensions, and cell lines. Whether working with tumor tissues, blood samples, or organ tissues, our advanced single-cell isolation technologies ensure efficient gene expression analysis while maintaining the integrity and accuracy of your samples.
Q2: How are specific cell types identified?
Through single-cell RNA sequencing, MtoZ Biolabs can accurately analyze gene expression profiles of individual cells. By comparing expression patterns to known marker genes or using data-driven clustering techniques, we can identify distinct cell types and subpopulations, revealing their specific functions within biological processes.
How to order?







