Single Cell Methylation Sequencing Service
- Cell Source: Samples can be sourced from various tissues, organs, or cell lines, as long as the cells are viable and not degraded.
- Cell Number: A minimum of 10,000 cells per experiment is required to ensure statistical validity and representativeness.
- DNA Quality: High-quality, purified DNA samples are essential for optimal sequencing performance.
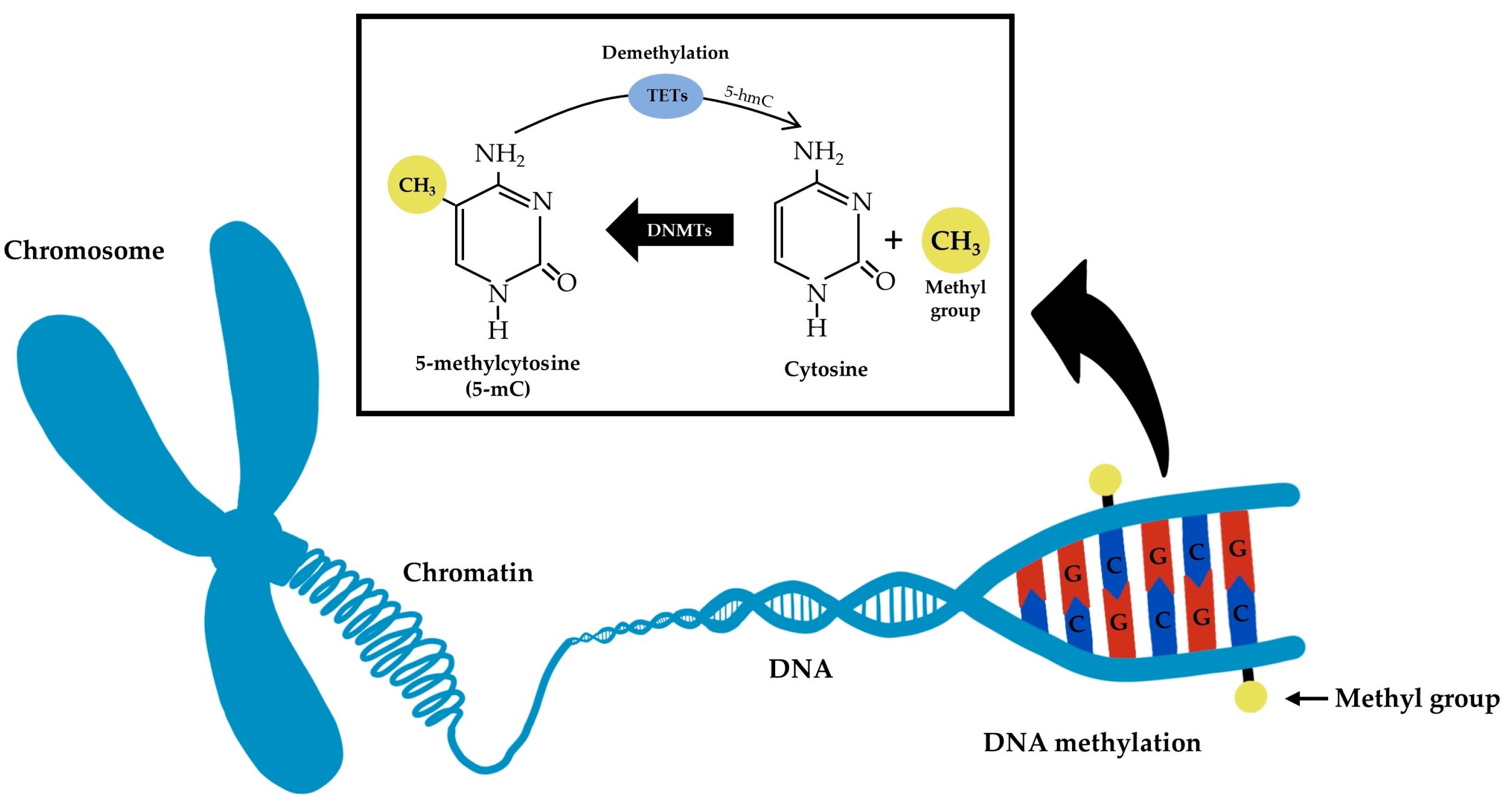
Single cell methylation sequencing is an innovative technology that enables comprehensive analysis of DNA methylation patterns at the single cell level. DNA methylation refers to the addition of a methyl (CH₃) group to the cytosine (C) residue in the DNA molecule, a key epigenetic modification. This modification does not alter the DNA sequence, but it can finely regulate gene expression. DNA methylation plays a crucial role in various biological processes in mammals, including genomic imprinting, X-chromosome inactivation, aging, and cancer progression.
Specifically, in oncology, aberrant DNA methylation is closely associated with the initiation and progression of cancer. Tumor cells often exhibit hypomethylation, leading to the overexpression of oncogenes, while hypermethylation of tumor suppressor genes may result in their inactivation. These methylation alterations have been identified as potential biomarkers for cancer diagnosis, prognosis, and personalized therapy. Therefore, analyzing DNA methylation at the single cell level can uncover early epigenetic changes in diseases such as cancer and offer insights for precision medicine.
Valente, A. et al. Nanomaterials. 2023.
Figure 1. Simplified Schematic Representation of DNA Methylation and Demethylation Molecular Mechanisms
MtoZ Biolabs offers high-precision, high-resolution Single Cell Methylation Sequencing Service using Whole Genome Bisulfite Sequencing (WGBS) technology, enabling clients to make groundbreaking advances in genomics and epigenetic research.
Technical Principles
The core of our Single Cell Methylation Sequencing Service is based on Whole Genome Bisulfite Sequencing (WGBS). This method utilizes bisulfite treatment to convert unmethylated cytosine (C) to uracil (U), while methylated cytosine (5-mC) remains unchanged. By applying this conversion, methylated and unmethylated sites can be distinguished after sequencing using alignment algorithms, generating a comprehensive methylation map across the entire genome. WGBS provides complete coverage of the genome and offers high sensitivity, making it ideal for single cell studies. Compared to traditional methylation chip or PCR techniques, WGBS provides superior sensitivity and broader coverage, capturing every methylation site in the genome.
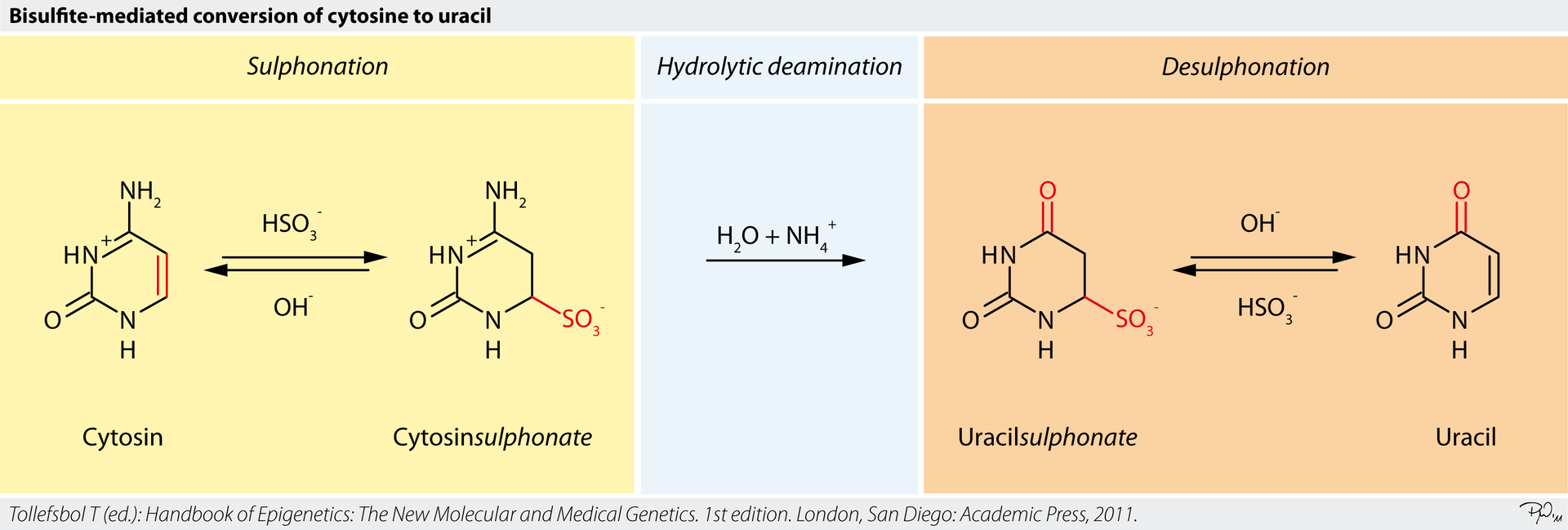
Figure 2. WGBS Chemical Basis: Bisulfite-Mediated Cytosine-to-Uracil Conversion
Analysis Workflow
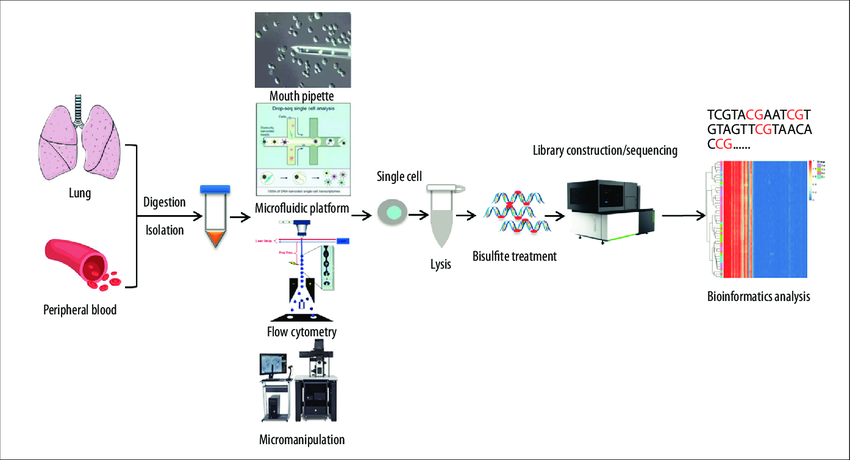
Our Single Cell Methylation Sequencing Service Process is streamlined and efficient, ensuring the highest quality experimental results:
1. Single cell Isolation
Single cells are isolated and captured using flow cytometry or microfluidic chip technology, ensuring the purity and accuracy of the data.
2. DNA Extraction and Bisulfite Treatment
DNA is extracted from single cells and treated with bisulfite, converting unmethylated cytosine to uracil, while methylated cytosine remains unchanged.
3. Library Construction and High-throughput Sequencing
Sequencing libraries are constructed and subjected to high-throughput whole-genome sequencing to obtain comprehensive single cell methylation data.
4. Data Analysis and Reporting
Bioinformatics tools are used to analyze the sequencing data and generate a detailed methylation map, allowing for in-depth interpretation of the research findings.
Men, S. et al. Med Sci Monit. 2020.
Service Advantages
· Single cell Resolution
Provides precise methylation data for individual cells, eliminating the heterogeneity of population-based data.
· Whole-genome Coverage
Delivers methylation data across the entire genome, including critical regions such as coding sequences, promoters, and enhancers.
· High Sensitivity and Accuracy
Utilizes state-of-the-art single cell methylation sequencing platforms to ensure high sensitivity and data accuracy.
· Tailored Services
Offers personalized experimental designs and data analysis to meet the unique needs of your research.
· One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
Sample Submission Suggestions
Proper sample preparation is essential for the success of the experiment and the accuracy of the data in our Single Cell Methylation Sequencing Service. We recommend the following guidelines:
If your samples require special treatments or preprocessing (such as activation or differentiation), please contact us in advance so that we can tailor an appropriate experimental plan.
MtoZ Biolabs provides high-quality Single Cell Methylation Sequencing Serviece based on cutting-edge WGBS technology, offering precise methylation data for a variety of research fields including genomics, oncology, immunology, and more. Let us help you advance your research by tailoring the optimal experimental plan. Contact us for a free consultation.
Case Study
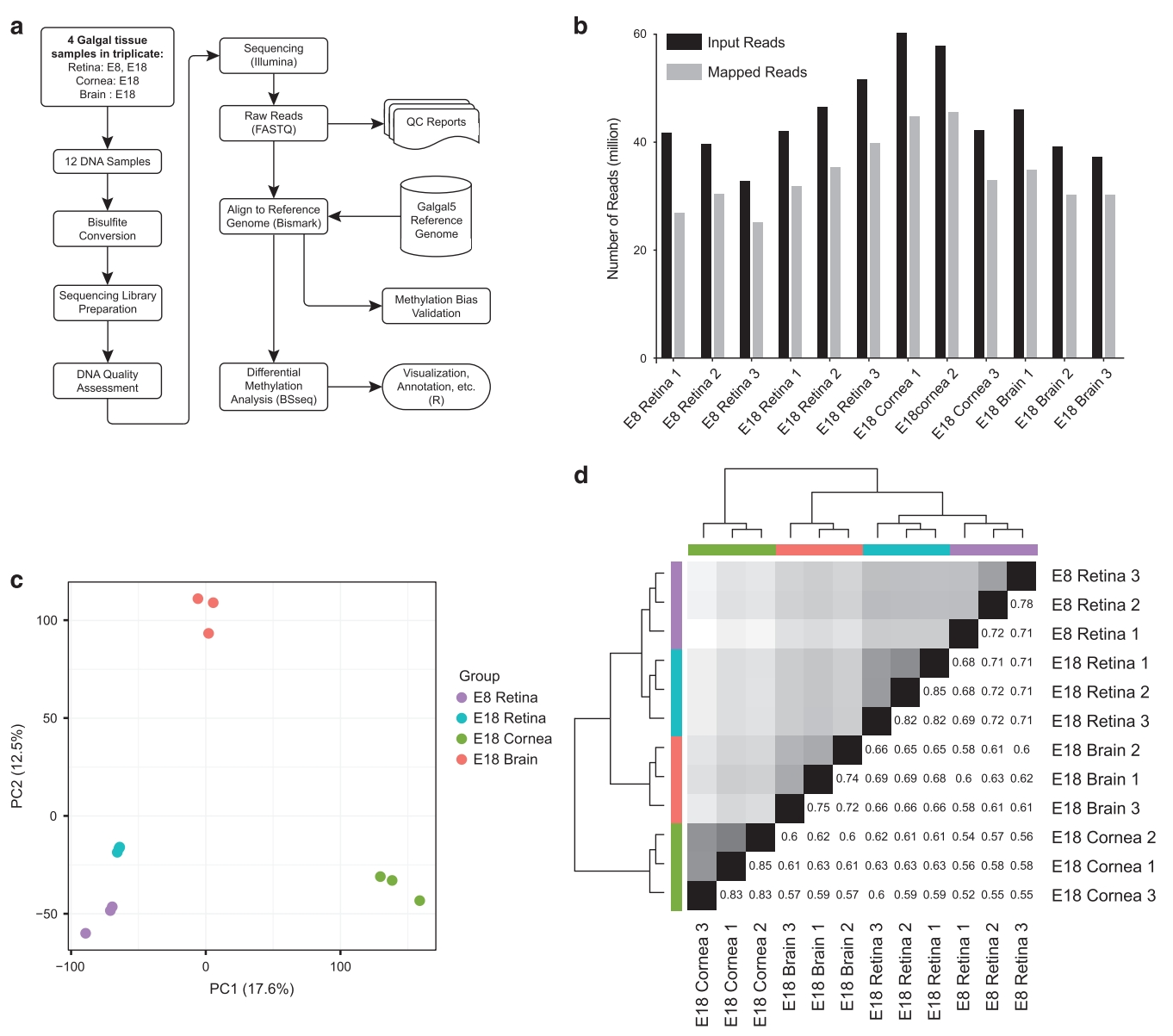
Case 1: Single Cell Methylation Sequencing Reveals Tissue-Specific Epigenetic Features in Chickens
In this study, single cell methylation sequencing was employed to investigate the methylation patterns in the retina, cornea, and brain tissues of chickens. The results highlighted key methylation regions associated with tissue-specific functions and gene regulation, underscoring the role of methylation in the regulation of the nervous and visual systems. This case demonstrates the application value of Single Cell Methylation Sequencing Service in epigenetic research.
Lee, I. et al. Sci. Data. 2017.
FAQ
Q1: Can single cell methylation sequencing be applied to all cancer types?
Yes, our Single Cell Methylation Sequencing Service can be used for various cancer types, particularly those with significant cellular heterogeneity, such as breast cancer, lung cancer, and hematologic malignancies.
Q2: How can single cell methylation sequencing be integrated with other single cell technologies?
Combining single cell methylation sequencing with other techniques like RNA sequencing and chromatin accessibility analysis provides a comprehensive view of cell states. This multi-omics approach enhances our understanding of the complex regulatory networks driving cancer.
How to order?







