SILAC Proteomics Analysis Service
The principle of Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) proteomics is based on the metabolic incorporation of stable isotope-labeled amino acids into the entire proteome. In SILAC, two cell populations are cultured in two different media: one contains "light" media with natural isotopic amino acids, while the other contains "heavy" media with stable isotope-labeled amino acids. After sufficient cell divisions, all proteins in the cells cultured in the "heavy" medium incorporate the heavy isotope-labeled amino acids. By analyzing the ratio of isotopically labeled peptides to unlabeled peptides using LC-MS/MS, the signal intensities of light and heavy amino acids in the cell culture are detected, enabling the relative quantification of their abundance in the mixture. SILAC proteomics analysis service provides quantitative protein analysis, offering precise relative quantification and identifying and quantifying relative differences in protein abundance. This approach does not require any chemical derivation or manipulation. Additionally, differentially treated samples can be combined at the level of intact cells or proteins, minimizing experimental error.
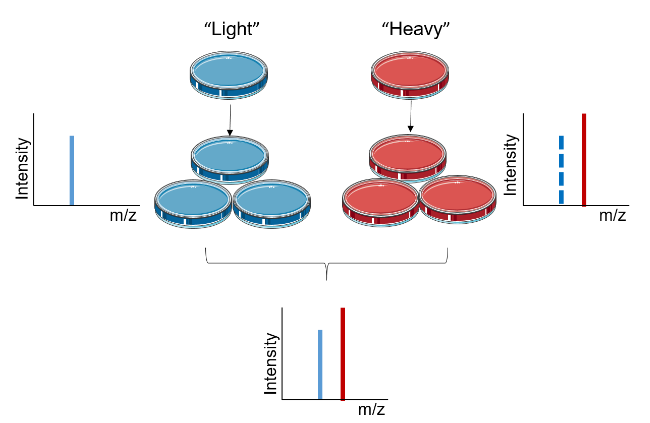
Chen, X. et al. Proteomics, 2015.
Figure 1. Experimental Principle of SILAC.
Services at MtoZ Biolabs
MtoZ Biolabs is committed to providing professional services such as quality control testing and project validation for the biotechnology, pharmaceutical, and medical device industries. MtoZ Biolabs provides SILAC proteomics analysis service and has established a complete proteomics platform, including Label-Free Quantitative Proteomics, TMT-Labeled Proteomics, DIA Quantitative Proteomics, 4D Quantitative Proteomics, PRM Targeted Protein Relative/Absolute Quantitation, and Metaproteomics. Additionally, we have established metabolomics and antibody sequencing platforms for comprehensive analytical support.
Service Advantages
1. Advanced Analytical Platform
MtoZ Biolabs offers a complete end-to-end workflow, including kit-based high-throughput sample preparation, automated fractionation, high-performance mass spectrometry acquisition, and professional data analysis and quality control platforms, ensuring reliable, fast, and precise analytical services.
2. Transparent Pricing
Our pricing is transparent, with no hidden or additional fees.
3. High-Quality After-Sales Service
From contract signing to sample receipt, experimental execution, and report delivery, if you have any questions about the report or experimental data within three years of completing the report, the after-sales service is online 24 hours a day to solve any problems related to the project at any time.
4. Customized Research Solutions
MtoZ Biolabs provides tailored services to address your unique research challenges and experimental requirements.
Applications
SILAC proteomics analysis service combines SILAC labeling with LC-MS/MS to analyze data, widely used to characterize quantitative protein differences between samples, study post-translational modifications (PTMs), and distinguish specific interacting proteins in protein-protein interaction (PPI) networks.
1. Expression Proteomics
SILAC provides an in vivo strategy to label proteins with different stable isotope forms of amino acids, enabling monitoring of quantitative differences in protein levels under various conditions. Moreover, SILAC proteomics analysis service can identify differentially expressed proteins in organelles, such as nuclei, nucleoli, or cellular insulin secretory granules.
2. Dynamic Changes in PTMs
For PTMomics analysis, SILAC-labeled peptides require fractionation and enrichment to improve PTM identification. Combined with MS technology, SILAC proteomics enables global and dynamic analysis of PTMs, including phosphorylation, acetylation, glycosylation, ubiquitination, and methylation.
3. PPI Studies
In PPI studies, protein complexes are immunoprecipitated from mixtures of SILAC-labeled cell lysates. SILAC can effectively differentiate specific interacting proteins from nonspecific background proteins. SILAC proteomics amalysis service is applicable for identifying specific interacting proteins in exogenous, endogenous, or inducible PPIs.
Case Study
1. SILAC-based quantitative proteomics can identify specific interacting proteins in exogenous PPIs, endogenous PPIs, or inducible PPIs.
When studying exogenous protein complexes (Figure 2A), wild-type cells or cells expressing affinity-tagged proteins are grown in either light or heavy media. Immunoaffinity purification of protein complexes is performed from a mixture of lysates obtained from the light and heavy cells. For endogenous protein complexes (Figure 2B), cells are grown in light or heavy media, and RNAi is used to knock down the target protein in one set of cells. Antibodies corresponding to the target protein are then used to immunoprecipitate the protein complexes from a mixture of lysates obtained from the light and heavy cells. In the case of inducible PPIs, protein complexes are induced through specific stimulation in cells grown in light or heavy media. Immunoaffinity purification is performed on the protein complexes from a mixture of lysates obtained from the light and heavy cells. After obtaining the protein complexes, the proteins are digested into peptides and analyzed using LC-MS/MS. Specific interacting proteins can be distinguished from nonspecific background proteins by their SILAC ratios.
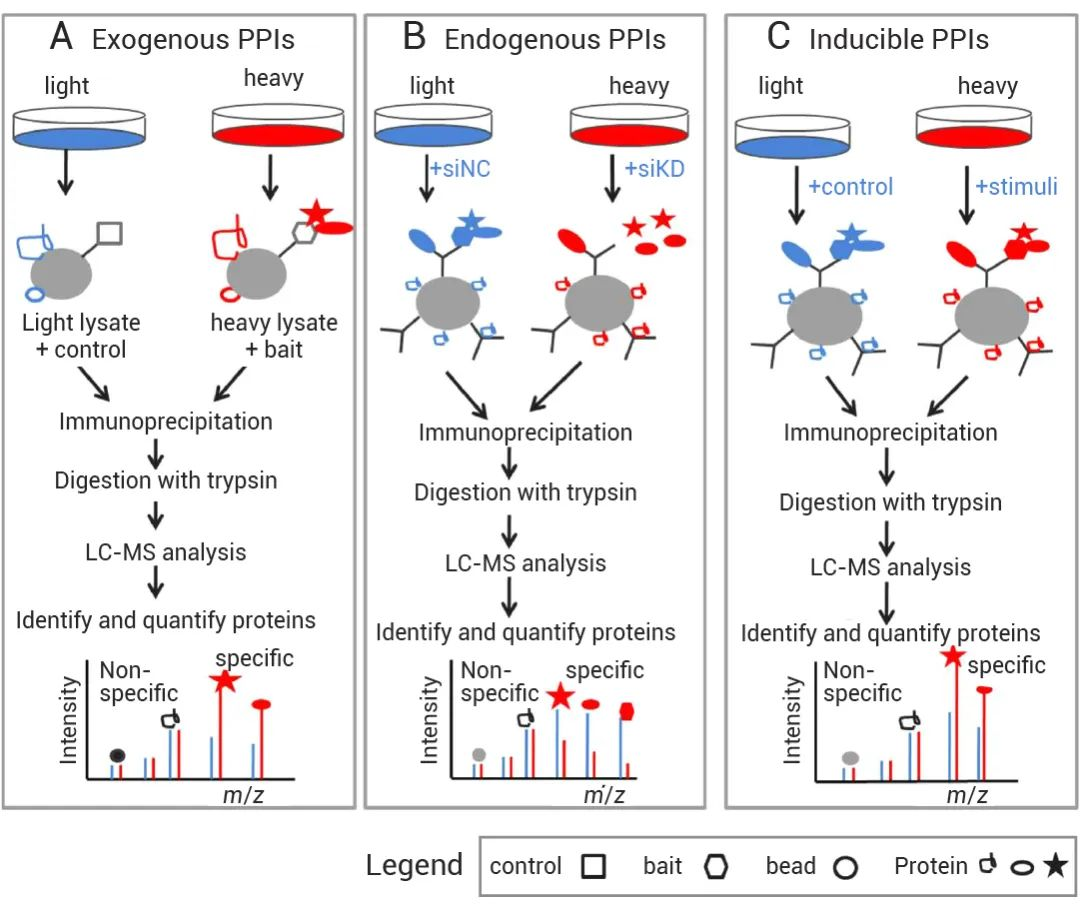
Chen, X. et al. Proteomics, 2015.
Figure 2. SILAC Quantitative Interaction Proteomics.
Deliverables
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Mass Spectrometry Images
4. Raw Data
5. Protein Differential Level Analysis
6. Bioinformatics Analysis
7. Customized Data Analysis
(1) Standardized Data Analysis Content: Protein identification and quality control analysis; Differential expression analysis; Comparative analysis across multiple groups, etc.
(2) Advanced Data Analysis Content: Functional analysis; Drug target annotation; Biomarker screening; Integration with clinical characterization for comprehensive insights, etc.
Note: For any specific requirements or personalized data analysis needs, please contact us.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Proteomics Service
How to order?







