Selenylation Proteomics Service
Selenylation Proteomics Service is a systematic proteomics service based on a high-resolution mass spectrometry platform. By combining chemical labeling and enrichment strategies, it enables the comprehensive identification, localization, and quantification of selenylation modification sites in proteins (e.g., Cys-SeH, Cys-Se-Cys, Cys-SeO). Integrated with bioinformatics analysis, this service also helps elucidate the physiological functions and pathological relevance of selenylation.
Qi Z. et al. Molecules. 2024.
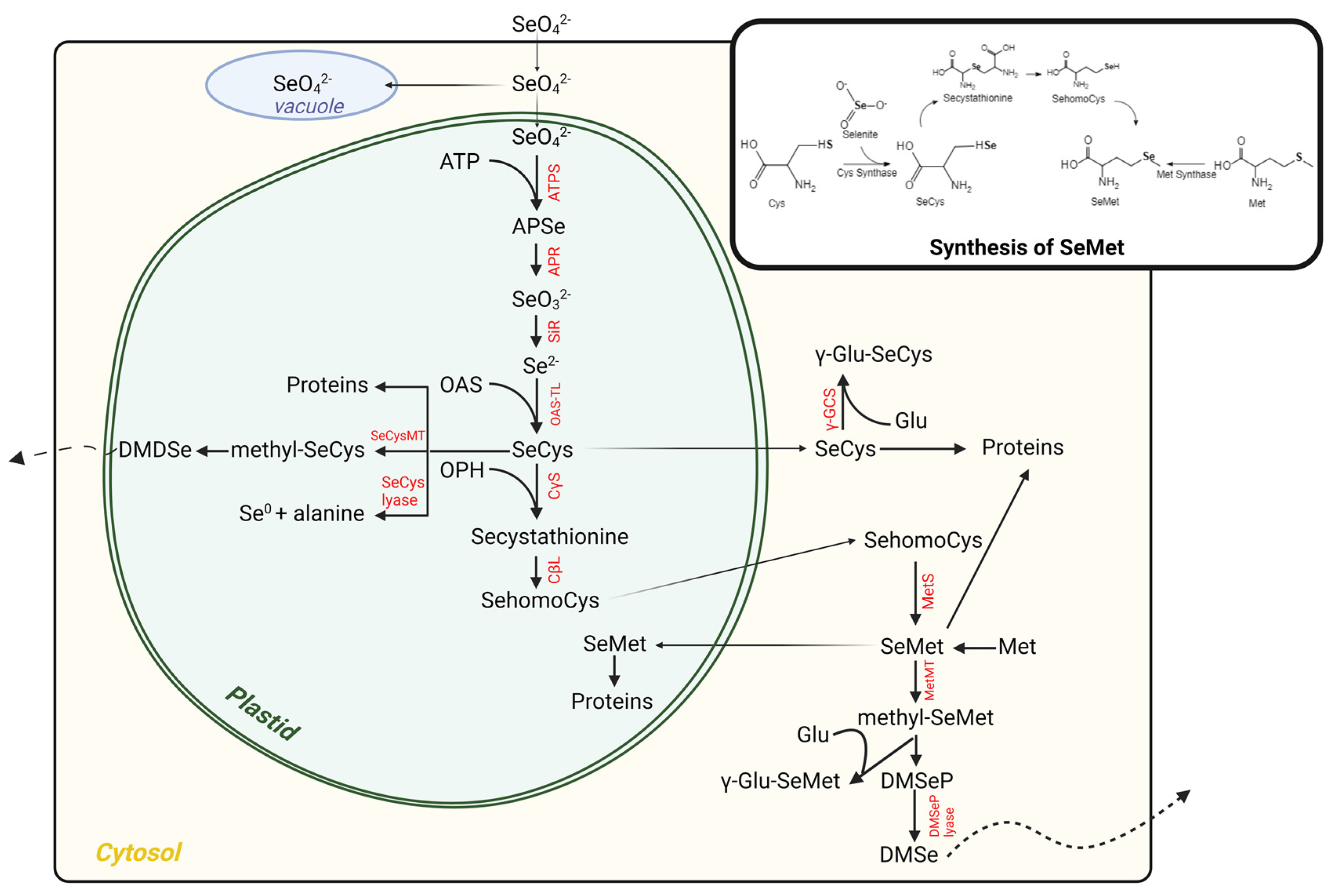
Figure 1. Schematic overview of Se metabolism in plants leading to the production of organic Se.
Selenium is one of the essential trace elements in the human body, involved in regulating key biological processes such as antioxidant defense, immune response, and apoptosis. Recent studies have shown that selenium exists not only in the form of selenocysteine (Sec) in proteins such as glutathione peroxidase (GPx), but also as a reversible covalent modification by forming Se-S, Se-N, or Se-O bonds with reactive protein residues (e.g., cysteine). This emerging type of post-translational modification, known as selenylation, has been demonstrated to play critical roles in redox regulation, antioxidant defense, cellular signal transduction, immune response, and tumorigenesis. Therefore, systematic investigation of selenylation modifications contributes to understanding the role of selenium in protein regulatory networks and promotes functional research and drug development targeting selenylation.
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs offers Selenylation Proteomics Service to comprehensively identify, localize, and quantify selenylation modification sites in proteins, construct associated functional regulatory networks, and support basic research, mechanistic studies, and drug screening efforts.
Analysis Workflow
The main workflow of Selenylation Proteomics Service is as follows:
1. Sample Preparation
Extract proteins from target tissues or cells, followed by reduction, alkylation, and enzymatic digestion.
2. Selenylation Labeling and Enrichment
Derivatize selenylated residues using specific chemical probes or tags, and purify modified peptides through affinity enrichment techniques.
3. Mass Spectrometry Analysis
Utilize high-resolution mass spectrometry and chromatography platforms to separate and detect peptides, capturing signal features of selenylated peptides.
4. Data Processing and Modification Identification
Apply specific mass shifts for selenylation in database searches to localize modified residues, and enhance specificity with characteristic fragment ions.
5. Functional Annotation and Result Delivery
Provide a list of modification sites, quantitative results, functional pathway annotations, and visual statistical charts.
Applications
Selenylation Proteomics Service is broadly applicable to the following research areas:
Biomedical Research
Investigate the role of selenylation in antioxidant defense, immune regulation, and apoptosis.
Disease Research
Explore potential biomarkers of selenylation in cancer, cardiovascular diseases, and neurodegenerative disorders.
Drug Development
Evaluate the impact of drugs on selenylation modifications to support drug screening and mechanism-of-action studies.
Environmental Monitoring
Monitor selenylation processes in environmental samples to assess selenium bioavailability and ecological effects.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Selenylation Proteomics Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Selenylation Proteomics Service data, providing clients with a comprehensive data report.
4. Comprehensive Workflow: End-to-end service from sample preparation to data analysis.
5. Customized Service: Tailored analysis plans and reporting formats based on client requirements.
FAQ
Q. How Can Sulfur Modifications be Distinguished from Selenium Modifications? Is There A Specific Recognition Mechanism?
To differentiate selenium from sulfur modifications, we use high-resolution LC-MS/MS platforms in combination with selenium-specific diagnostic ions (e.g., m/z ≈ 79, 81, 95) and characteristic isotopic distribution patterns. Naturally occurring selenium isotopes (78/80/82) are also utilized to strengthen specificity. For potentially overlapping S-modifications (such as disulfide bonds or S-nitrosylation), we further validate via retention time comparison of isomeric peptides, modification-specific enzymatic digestion, or manual spectrum review. This multi-layered strategy ensures high-confidence identification of Se modifications and minimizes the risk of false positives and misannotations.
Q. Can Se Modification Levels be Quantitatively Compared Across Different Treatment Groups? How Accurate is the Quantification?
Yes. MtoZ Biolabs supports both label-free quantification and TMT/iTRAQ labeling strategies for comparative analysis of selenium modifications across multiple conditions. Through standardized sample preparation protocols, internal calibration standards, and cross-batch normalization strategies, we ensure high precision and reproducibility in quantifying Se-modified peptides across samples. We also provide statistical significance testing and data visualization to help clients identify key differentially modified sites.
Case Study
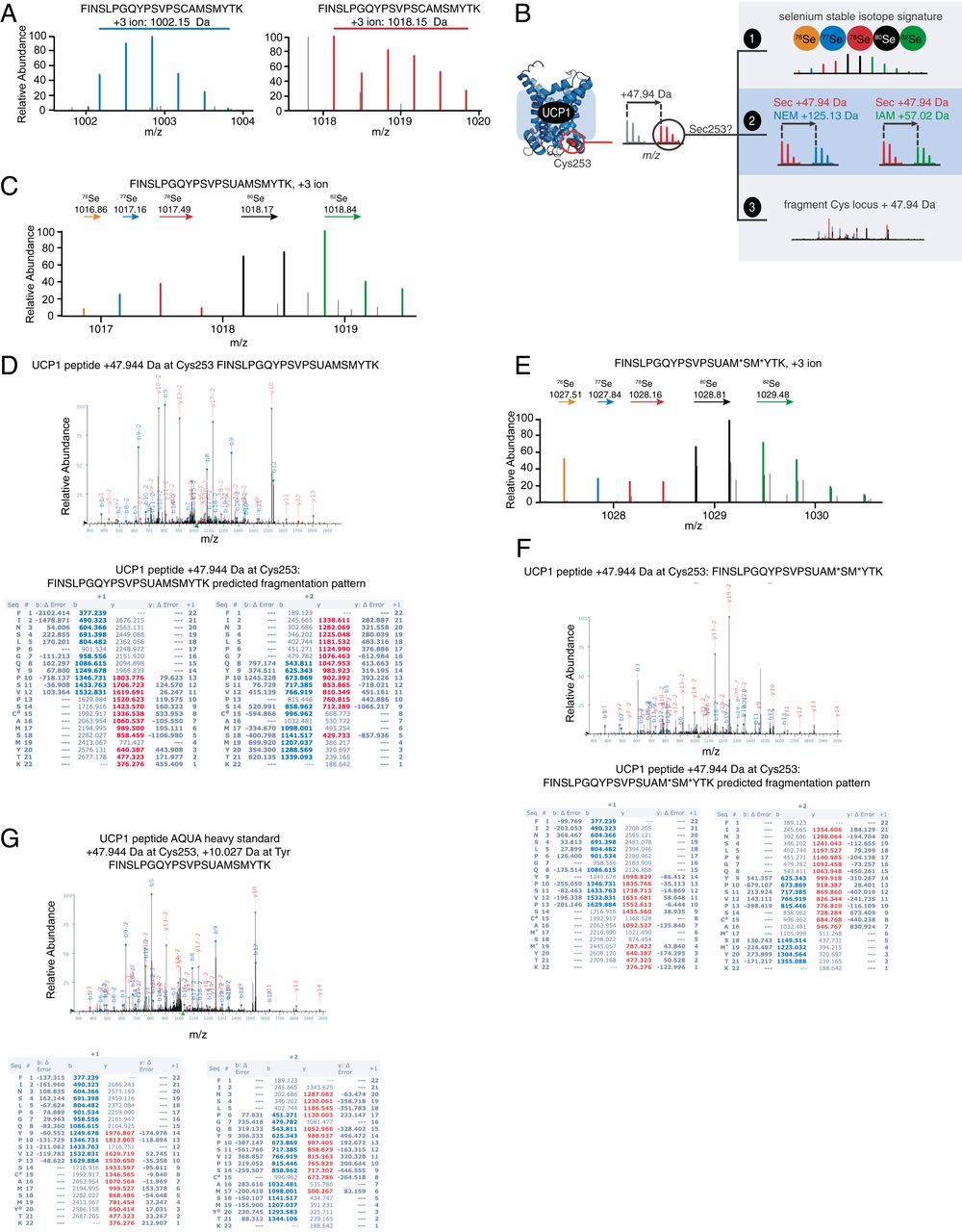
This study systematically analyzed non-canonical selenium incorporation in proteins from mammalian tissues. Using high-resolution mass spectrometry combined with selenium isotope-specific detection, the researchers discovered that selenium can be incorporated into several metabolism-related proteins—even in the absence of UGA codons and SECIS elements—in the form of selenocysteine or selenomethionine. These proteins include thermogenesis-regulating factors such as UCP1. Functional validation further showed that dietary selenium supplementation enhanced selenylation levels of UCP1, promoted thermogenic activity in brown adipose tissue, and attenuated high-fat diet–induced obesity. This study is the first to confirm in vivo inducible protein selenylation and reveals its important role in redox sensitivity regulation and energy metabolism.
Jedrychowski MP. et al. Proc. Natl Acad Sci USA. 2020.
Figure 2. Selenocysteine incorporation into 253 locus of UCP1.
How to order?







