S-nitrosylation Proteomics Service
S-nitrosylation Proteomics Service is a proteomics-based service designed to systematically identify, quantify, and functionally annotate S-nitrosylation modification sites, their abundance, and dynamic changes under different conditions in proteins.
S-nitrosylation is an important post-translational modification (PTM), referring to the covalent attachment of a nitric oxide (NO) group to the thiol (–SH) side chain of cysteine residues in proteins, forming an S-nitrosothiol (S–NO) bond. As a reversible redox-based modification, S-nitrosylation is recognized as a key regulatory mechanism in NO signaling. It plays broad roles in physiological and pathological processes including cardiovascular function, immune response, neuronal signaling, tumor development, and oxidative stress regulation.
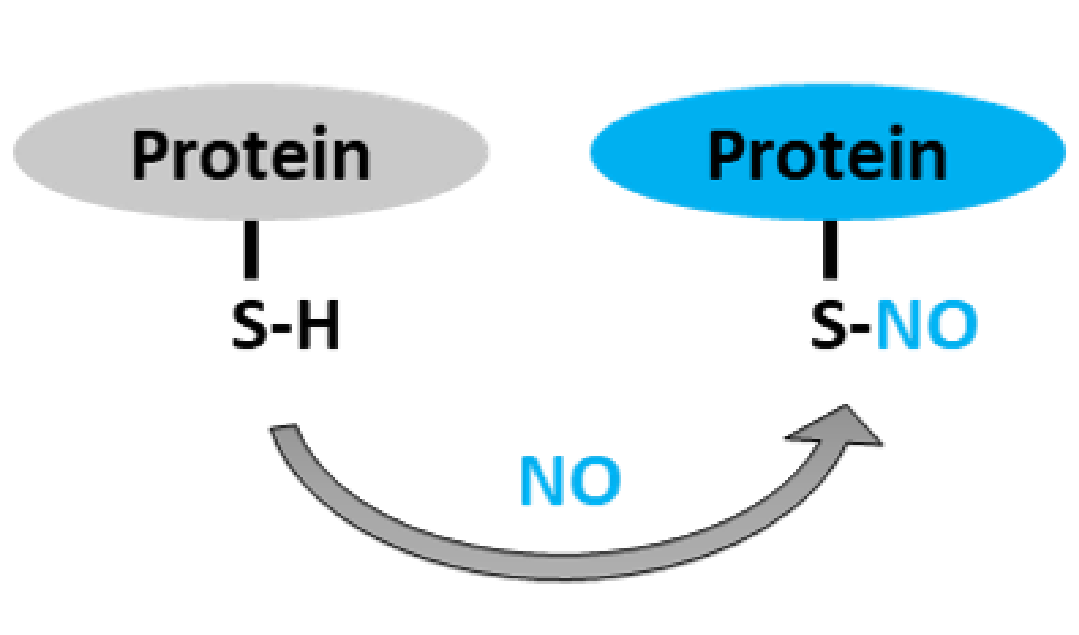
Sharma V. et al. Int J Mol Sci. 2021.
Unlike high-abundance modifications such as phosphorylation or acetylation, S-nitrosylation is characterized by its dynamic nature, low abundance, and strong condition dependency, making its site-specific detection particularly challenging. In recent years, the development of specific chemical labeling methods, enrichment strategies, and high-resolution mass spectrometry has positioned S-nitrosylation proteomics as a core approach for investigating NO-mediated signal regulation.
Leveraging advanced mass spectrometry and chromatography platforms, MtoZ Biolabs offers S-nitrosylation Proteomics Service with high sensitivity and specificity for the identification of S-nitrosylation sites in proteins. The service supports accurate quantification of site-specific changes under various treatments or pathological conditions, providing reliable data for elucidating NO-mediated signaling pathways and regulatory mechanisms.
Analysis Workflow
The main workflow of the S-nitrosylation Proteomics Service is as follows:
1. Sample Preparation
Extract total proteins from cells, tissues, or body fluids, followed by pretreatment with reducing agents and blocking of free thiol groups.
2. Selective Reduction and Labeling of S-NO Bonds
Use ascorbate to selectively reduce S–NO bonds, then perform thiol-specific labeling with biotin or TMT tags.
3. Enrichment of Modified Peptides
Enrich S-nitrosylated proteins/peptides using streptavidin magnetic beads or TMT-based pull-down strategies.
4. Mass Spectrometry Acquisition and Data Analysis
Perform LC-MS/MS to identify modified peptides, localize modification sites, and compare modification level differences.
5. Functional Annotation and Pathway Analysis
Deliver annotated modification spectra, site-level quantification heatmaps, and GO/KEGG pathway annotations.
Applications
S-nitrosylation proteomics has broad applications across multiple research areas, including but not limited to:
Mechanistic Studies of NO Signaling Pathways
Reveal how NO regulates the activity, stability, and interactions of key signaling proteins through S-nitrosylation.
Cardiovascular Disease Research
Compare S–NO modification profiles of myocardial proteins under conditions such as ischemia/reperfusion or inflammation.
Neurodegenerative Disease Research
Identify S–NO regulatory targets related to diseases such as Parkinson’s and Alzheimer’s.
Pathogen Infection and Immune Evasion Mechanism Studies
Analyze how pathogenic proteins respond to host NO-mediated stress via S-nitrosylation.
Drug Mechanism of Action Studies
Evaluate how drugs modulate S-nitrosylation patterns to assist in target identification and functional mechanism research.
FAQ
Q. Is There A Problem of Non-Specific Reduction in the Ascorbic Acid Reduction Method?
Yes. Although ascorbate is widely used in traditional methods such as the Biotin-Switch technique to efficiently reduce S–NO bonds, it may also reduce other modifications like S-glutathionylation or disulfide bonds under certain conditions, leading to non-specific signals. To improve confidence in site identification, MtoZ Biolabs recommends the following: include a non-reducing control group (minus-AA control) for background correction; use optimized reduction conditions; validate specific sites through point mutations, chemical inhibition, or enzymatic methods; and apply high-resolution mass spectrometry with advanced fragmentation methods (e.g., EThcD) to improve peptide localization accuracy.
Q. Can Sample Processing Balance the Stability of S-Nitrosylation and Protein Integrity?
Yes. Given the extreme instability of S–NO bonds, we perform standardized procedures during sample preparation—including light protection, low temperature (4°C), mildly acidic buffer conditions, and rapid blocking of free thiols—to preserve S-nitrosylation from oxidation or conversion. In addition, we offer optimized protocols tailored for tissues, cells, and body fluids to ensure that S–NO modifications are retained while maintaining protein integrity and downstream analytical quality.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study compared the protein S-nitrosylation patterns of Escherichia coli under different lifestyles (planktonic vs. biofilm) and oxygen conditions (aerobic vs. anaerobic). By combining the biotin-switch technique with stable isotope labeling (SILAC), the researchers identified numerous proteins that undergo biofilm-specific S-nitrosylation, particularly those involved in redox homeostasis and amino acid biosynthesis, such as OxyR, KatG, and GltD. Functional validation experiments demonstrated that S-nitrosylation of these proteins plays a critical role in regulating E. coli motility, biofilm maturation, and resistance to oxidative stress.
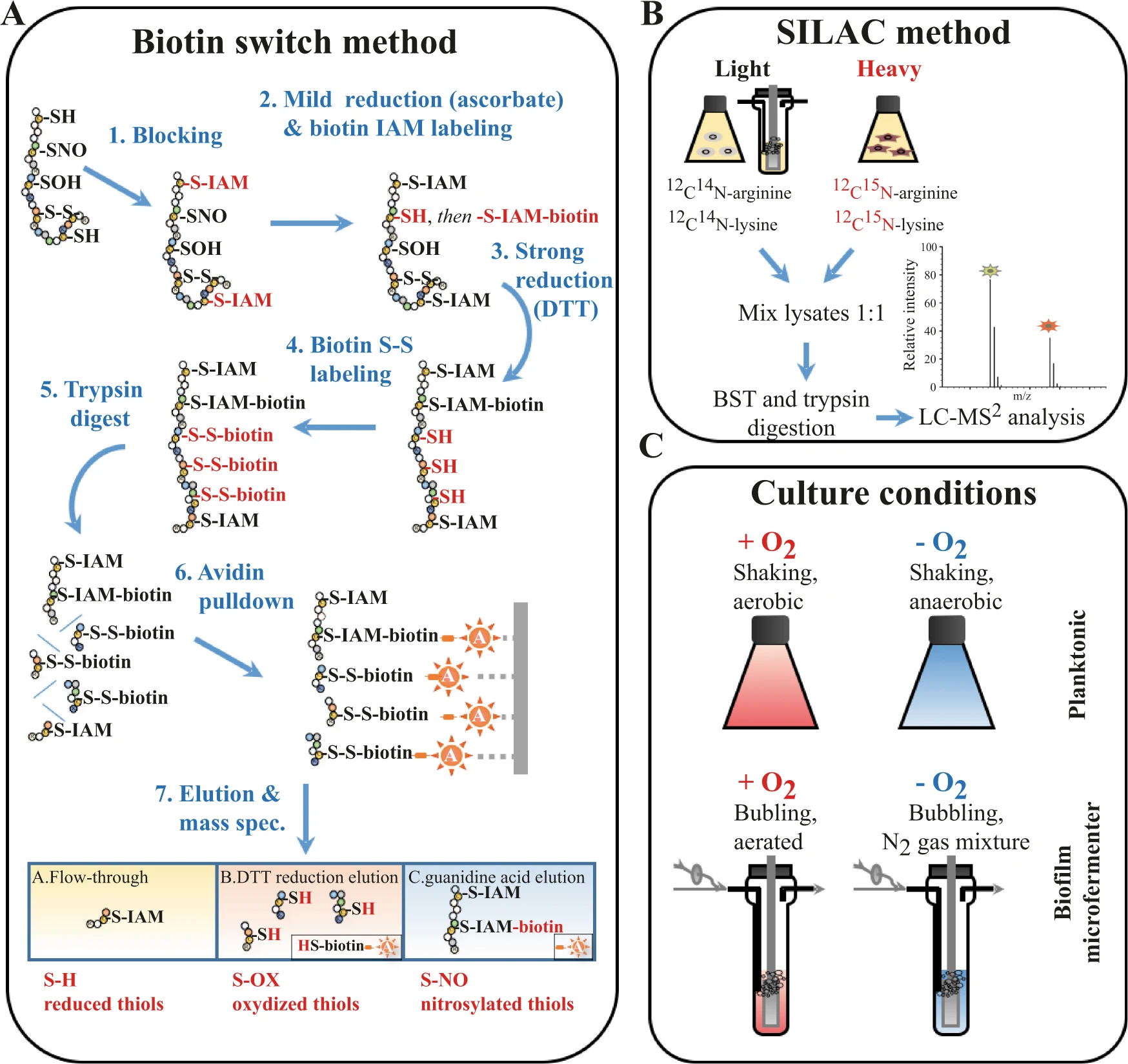
Barraud N. et al. npj Biofilms Microbiomes. 2021.
How to order?







