Quantitative Approaches and Strategies in Chemical Proteomics
-
Custom design and synthesis of reaction-specific chemical probes
-
Automated platforms for sample enrichment and enzymatic digestion
-
Versatile quantitative strategies (TMT, Label-Free, SILAC)
-
High-resolution Orbitrap mass spectrometry systems (Exploris, Fusion Lumos)
-
A dedicated bioinformatics team for functional protein profiling and data visualization
Proteomics traditionally emphasizes variations in protein expression levels; however, this approach falls short in comprehensively capturing the active or functional states of proteins. For instance, a protein may retain a constant expression level while undergoing substantial alterations in activity due to post-translational or covalent modifications. Quantitative analysis in chemical proteomics offers a novel strategy to overcome this limitation. By employing chemically reactive and target-specific probes, researchers can selectively label, enrich, and quantify protein populations in their active states, thereby uncovering the true functional roles of proteins in dynamic biological processes.
Core Principles of Chemical Proteomics
1. Activity-Based Probes (ABPs)
ABPs serve as the fundamental tools in chemical proteomics and are typically composed of three key components:
(1) Recognition group: designed to specifically bind the active site of the target protein
(2) Reactive group: forms a covalent bond with the protein, ensuring stable attachment
(3) Tagging group: such as biotin, fluorophores, or isotope tags, which facilitate downstream enrichment and detection
This modular design enables ABPs to achieve high selectivity in labeling active proteins within complex biological samples, thereby circumventing the reliance on protein expression levels in conventional proteomics.
2. Click Chemistry and Enrichment Strategies
To enhance both quantitative accuracy and sensitivity, modern chemical proteomics often incorporates Click Chemistry for rapid post-labeling conjugation. This is typically followed by affinity-based enrichment strategies using streptavidin-coated magnetic beads or specific antibodies, enabling efficient recovery of low-abundance active proteins.
Overview of Quantitative Approaches in Chemical Proteomics: From Probe Design to Mass Spectrometry Data
Quantitative analysis in chemical proteomics generally involves the following key stages:
1. Probe Design and Sample Preparation
Probe selection is dictated by the enzymatic or functional class of the target proteins. For example:
(1) Serine hydrolases: fluorophosphonate-based probes are employed
(2) Cysteine proteases: typically targeted using alkylating groups such as acetylenyl amides
(3) Protein tyrosine phosphatases: require tailored probes containing para-substituted alkylphenylphosphonic acid moieties
Probes are introduced into cell lysates or live cell cultures to react selectively with their respective protein targets.
2. Enrichment of Probe–Protein Complexes
Following probe–target binding, Click Chemistry is employed to attach biotin or fluorescent labels. The resulting complexes are enriched using streptavidin magnetic beads. At this stage, MtoZ Biolabs utilizes a high-throughput automated enrichment platform, significantly enhancing sample processing consistency and recovery efficiency.
3. Proteolytic Digestion and Quantitative Labeling
The enriched proteins are digested with trypsin. Quantitative comparison across experimental conditions can then be achieved through isotopic labeling methods such as TMT or iTRAQ, or alternatively, label-free quantification approaches can be employed for specific analytical systems.
4. LC-MS/MS Analysis and Data Interpretation
High-resolution mass spectrometry (e.g., Orbitrap Exploris 480) is used to perform both qualitative and quantitative peptide analysis. The resulting data are processed using software such as MaxQuant, Proteome Discoverer, or proprietary algorithms to evaluate changes in protein activity across conditions.
Constructing Optimal Analytical Strategies Based on Sample Types
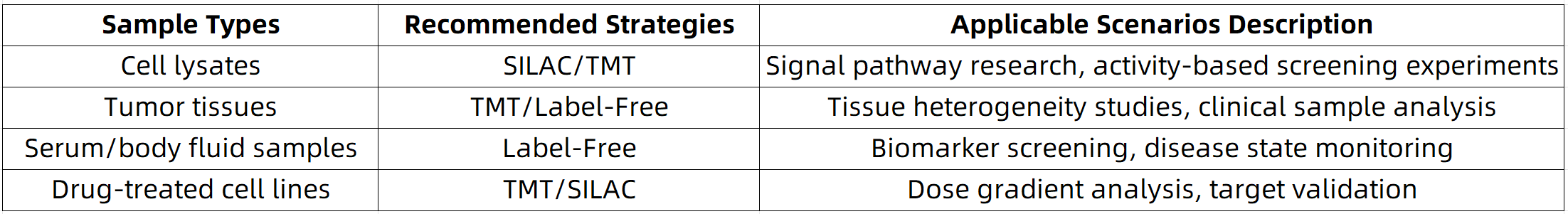
Advantages of MtoZ Biolabs: From probe optimization to high-throughput quantification
MtoZ Biolabs offers comprehensive chemical proteomics solutions, including but not limited to:
By leveraging these strategies, we assist researchers in identifying functionally relevant proteins within complex biological matrices, thereby accelerating target discovery, biomarker identification, and mechanistic investigations. As biomedical science advances from transcriptomics to functional proteomics, chemical proteomics is emerging as an essential technology. With precise chemical probe labeling and high-throughput mass spectrometry, researchers can pinpoint key proteins in their active states directly from native samples, facilitating a seamless transition from fundamental research to therapeutic innovation.
For researchers investigating protein activity, target identification, or mechanistic pathways, MtoZ Biolabs provides specialized technical support and tailored analytical services to advance your scientific goals.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







