PTM Quantitative Analysis Service
PTM Quantitative Analysis Service is designed to quantitatively detect post-translational modifications (PTMs) in proteins, revealing their spatiotemporal distribution, dynamic changes, and roles in biological processes. PTMs are widely involved in cellular signal transduction, gene expression regulation, immune response, and metabolic processes. To fully understand the role of PTMs in biological processes, it is crucial to accurately quantify the changes in various PTMs. Mass spectrometry is one of the commonly used techniques for PTM quantitative analysis. It can directly calculate the relative abundance of PTMs in different samples by measuring the peak area or intensity of specific modified peptides. Additionally, PTM quantification can be performed through combination with label-based quantification techniques (such as TMT, iTRAQ) and affinity capture methods (such as immunoprecipitation) for relative or absolute quantification. Leveraging advanced mass spectrometry platforms and a variety of quantitative strategies, MtoZ Biolabs offers PTM Quantitative Analysis Service that accurately measures PTM abundance changes in proteins. Whether for the quantification of single modifications or integrated analysis of multiple modifications, we provide research support to help scientists explore the dynamic changes and biological significance of post-translational modifications.
Heitel P. Molecules. 2023.
Analysis Workflow
1. Sample Preparation
Extract proteins and perform enzymatic digestion.
2. PTM Enrichment and Labeling
Depending on the analysis objective, apply specific PTM enrichment methods to isolate modified peptides from the sample. If quantification is required, label the samples using appropriate tagging techniques.
3. Mass Spectrometry Analysis
Perform protein analysis using high-resolution LC-MS/MS to obtain mass-to-charge ratio (m/z) information of modified peptides.
4. Data Analysis and Reporting
Process mass spectrometry data using specialized software to identify modification sites and conduct quantitative analysis. Integrate bioinformatics analysis to generate a comprehensive PTM quantitative analysis report.
Applications
Cell Signaling Transduction Research
Quantitative analysis of PTM changes in signaling pathways helps reveal the regulatory roles of proteins in signal transmission, providing insights into the molecular mechanisms of cellular responses to external stimuli.
Disease Mechanism Research and Biomarker Discovery
By comparing PTM differences between normal and disease states, disease-associated biomarkers can be identified.
Protein Function Regulation Analysis
Investigate the regulatory effects of PTMs on protein activity, stability, localization, and interactions to elucidate protein functions and mechanisms within cells.
Drug Development and Mechanism of Action Studies
PTM quantitative analysis aids in assessing the impact of drugs on target protein modifications. By analyzing drug-induced changes in protein modifications, the underlying mechanisms of drug action and its regulatory effects on target proteins can be elucidated.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced PTM Quantitative Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all PTM Quantitative Analysis Service data, providing clients with a comprehensive data report.
FAQ
Q. TMT/iTRAQ vs. Label-Free vs. SILAC, How to Choose the Optimal PTM Quantification Method?
TMT/iTRAQ (Multiplexed Label-Based Quantification): Suitable for multi-sample quantification, allowing PTM changes to be compared across multiple conditions within the same experiment. However, it may lead to ratio compression, potentially affecting quantification accuracy.
Label-Free Quantification (LFQ): Does not require labeling and is ideal for large-scale PTM analysis. While it has higher quantification variability, it is particularly useful for discovering novel PTMs.
SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture): Provides high-precision PTM quantification and is well-suited for cell culture samples, but it is not applicable to tissue or biofluid samples.
Q. How to Improve PTM Site Identification Accuracy and Avoid False Positives?
1. Optimize Database Search Strategy: Set FDR ≤ 1% to control the false positive rate and use Delta Score or Ascore (≥13) to enhance confidence in PTM site identification.
2. Choose an Appropriate Fragmentation Mode: Phosphorylation benefits from HCD (Higher-energy Collisional Dissociation), while glycosylation is better analyzed using ETD (Electron Transfer Dissociation) to minimize modification loss.
3. Use Characteristic Fragment Ions for PTM Verification: Identify PTM types based on specific fragment ions, such as phosphorylation (97.98 Da), glycosylation (204 Da), and ubiquitination (114 Da).
4. Biological Validation: Confirm PTM sites using site-directed mutagenesis (S/T/Y → A/E) and Western blot with modification-specific antibodies.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study analyzed the dynamic changes in phosphorylated proteins within tumor cells before and after Rapamycin treatment to elucidate its anti-tumor mechanism. The results showed that Rapamycin significantly inhibits the phosphorylation of STAT3 (Signal Transducer and Activator of Transcription 3) at Tyr705, leading to reduced STAT3 activity. Consequently, the expression of its downstream target gene c-Myc is downregulated, thereby suppressing tumor cell proliferation and survival.
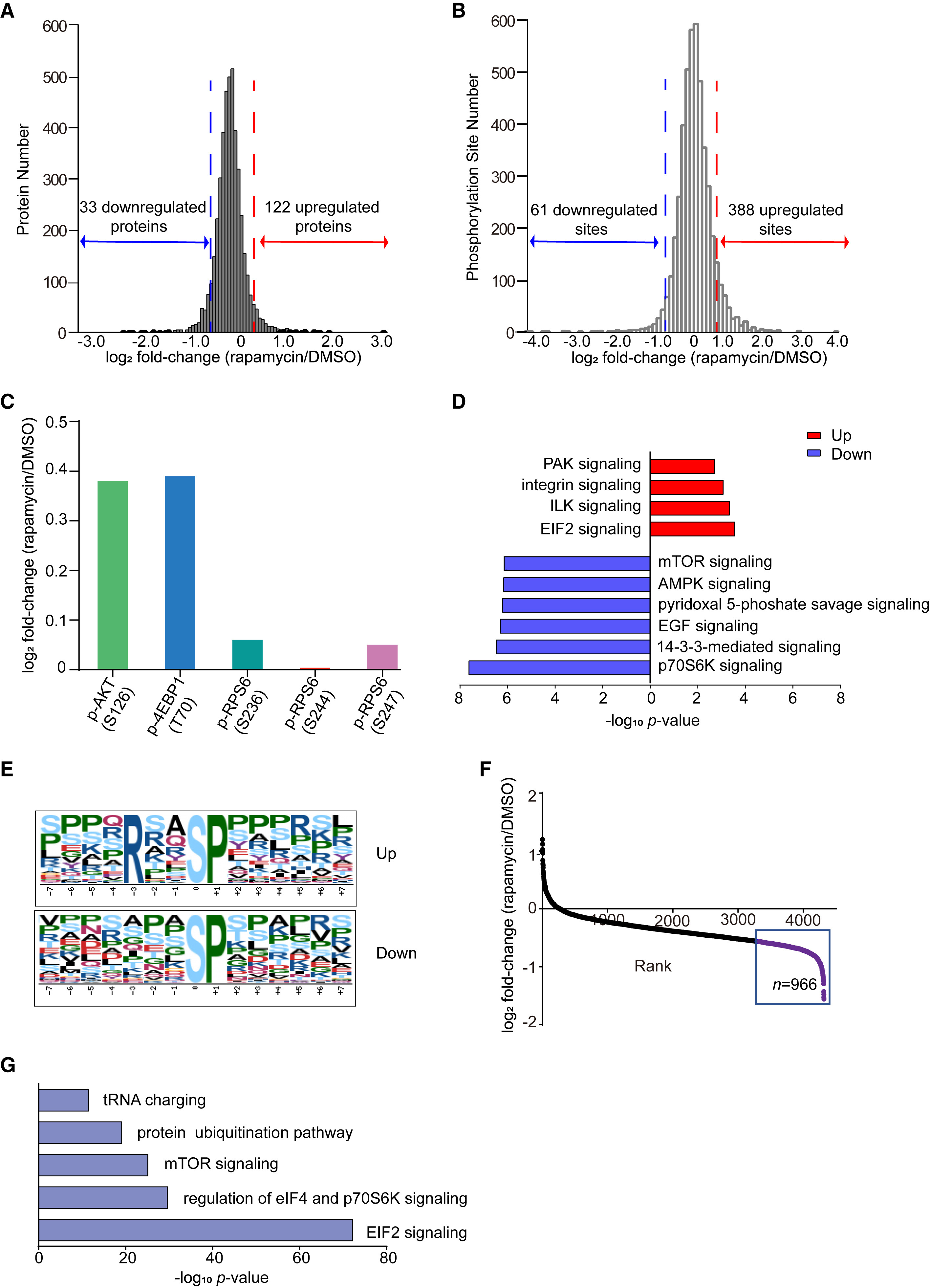
Sun L. et al. Cell chemical biology. 2022.
How to order?







