Proteomics Pathway Analysis Service
Proteomics pathway analysis is a critical branch of bioinformatics that integrates proteomics and computational analysis to elucidate the functions, interactions, and roles of proteins in specific biological processes. The proteome represents the complete set of proteins expressed by a cell, tissue, or organism under specific temporal and spatial conditions. Pathway analysis involves investigating the interactions and regulatory relationships among proteins, genes, and other molecules to construct biological pathways, thereby providing insights into fundamental life processes. Within living organisms, proteins function cooperatively within complex interaction networks to execute a wide range of biological functions.
Proteomics pathway analysis service facilitates the identification of key regulatory nodes by studying protein expression, modifications, localization, and interactions. This approach enhances our understanding of the molecular mechanisms underlying disease onset and progression, providing new insights for disease diagnosis, treatment, and prevention.
Services at MtoZ Biolabs
Leveraging state-of-the-art instrumentation, MtoZ Biolabs has established an advanced proteomics pathway analysis service platform, offering a comprehensive one-stop analytical service covering the following aspects:
1. Sample Preparation and Protein Extraction
First, proteins are extracted from cells, tissues, or organisms. This process may involve centrifugation, filtration, and other methods to isolate proteins.
2. Protein Separation and Identification
The extracted proteins are separated using electrophoresis (such as SDS-PAGE), chromatography techniques, or mass spectrometry (MS). Mass spectrometry is particularly important as it provides information on protein mass and sequence, allowing for protein identification and the analysis of post-translational modification sites.
3. Quantitative Analysis
Quantitative analysis of protein expression levels is typically performed using mass spectrometry techniques, such as labeled quantification (e.g., isotope labeling) or label-free quantification methods (e.g., label-free quantification).
4. Bioinformatics Analysis
Various software and algorithms are used to analyze the obtained data to identify expression patterns, protein interactions, and pathway activation. This includes protein identification, enrichment analysis, network modeling, and pathway mapping.
5. Pathway Analysis
Protein data is mapped onto known biological pathways to identify active pathways or biological processes. This helps in understanding the molecular mechanisms occurring in biological samples, as well as changes in disease states or treatment responses. Commonly used databases include KEGG, Reactome, and WikiPathways.
6. Integration and Interpretation
Proteomics data is integrated with other omics data, such as genomics, transcriptomics, and metabolomics, to gain system-level biological insights. This helps in uncovering the mechanisms of complex diseases, identifying biomarkers, and discovering potential therapeutic targets.
Service Advantages
1. Advanced Analytical Platform
MtoZ Biolabs has established a proteomics pathway analysis service platform with a comprehensive protein database, enabling accurate, complete, and thorough database annotation. Our platform facilitates in-depth data mining, comparative analysis of species, gene composition, functional attributes, and metabolic pathway differences across samples. This provides valuable reference information for health research, disease prevention, and biomedical studies.
2. Transparent Pricing
Our pricing is transparent, with no hidden or additional fees.
3. Customized Research Solutions
MtoZ Biolabs offers tailored services to address unique research questions and experimental requirements.
Applications
1. Disease Mechanism Research
By analyzing the distribution of differentially expressed proteins within biological pathways, researchers can uncover key molecular mechanisms underlying disease onset, progression, and treatment responses, facilitating the discovery of novel therapeutic targets.
2. Drug Mechanism Exploration
Proteomics pathway analysis service can help elucidate how drugs influence specific biological pathways or signaling networks, optimizing drug design and predicting potential side effects.
3. Biomarker Discovery
Integrating pathway analysis with differential protein expression profiling enables the identification of key proteins and potential biomarkers associated with disease diagnosis and prognosis.
4. Functional Annotation and Data Mining
Proteomics pathway analysis provides biological context to differential proteins, clarifying their roles in metabolism, signaling pathways, cell cycle regulation, and other biological processes.
5. Multi-Omics Integration
By integrating genomics, transcriptomics, and other multi-omics data, proteomics pathway analysis service provides systematic insights to support the study of complex biological systems.
Case Study
1. Changes in the susceptibility and resilience of hippocampal proteome to depression or anxiety in a rat model with chronic mild stress
This study conducted quantitative proteomic analysis on different groups of model rats, including the control group, depression-susceptible group, anxiety-susceptible group, and resilient group. Differentially expressed proteins were identified and subjected to pathway analysis. The results showed that these differentially expressed proteins were primarily involved in metabolic processes such as the tricarboxylic acid (TCA) cycle, carbon and pyruvate metabolism, oxidative phosphorylation, and other key metabolic pathways. By integrating both wet-lab and computational analyses, the identified candidate biomarkers revealed potential protein targets involved in various metabolic pathways. This study provides valuable insights into the similarities and differences in the pathophysiological mechanisms underlying stress-induced depression or anxiety and stress resilience.
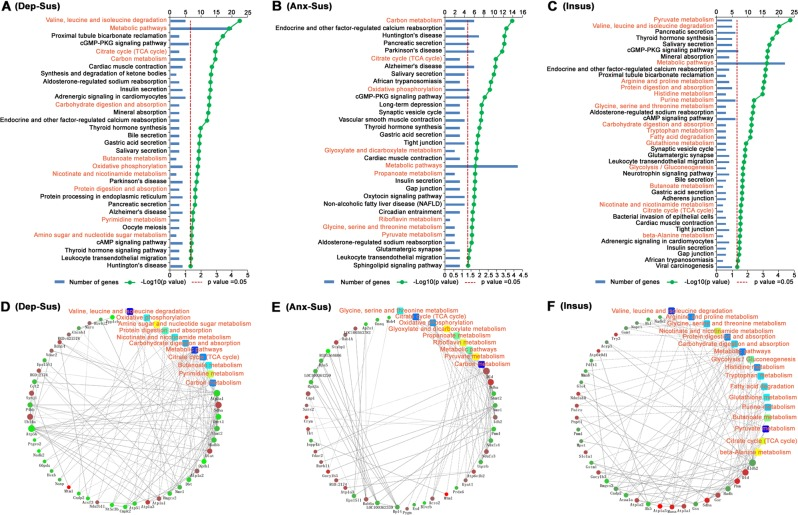
Tang, M. et al. Translational Psychiatry, 2019.
Figure 1. KEGG Pathway Enrichment and Protein-Protein Interaction (PPI) Network of Differentially Expressed Proteins.
2. HIF1A Attenuates Compression-Induced Apoptosis of Nucleus Pulposus Stem Cells by Upregulating Autophagy
In the study of HIF1A in relieving the apoptosis of nuclear pulposus derived stem cells (NPSCs) induced by compression, the researchers deeply analyzed the mechanism of HIF1A by combining with the Label-Free quantitative protein group technique. By comparing the differences between the compression+hypoxia group (CMP+HX) and the compression (CMP) group, they conducted various functional analysis on the different proteins to explore the potential pathway mechanism. The results revealed that the upregulated proteins in the CMP+HX group were significantly enriched in the autophagy process and HIF1A signaling pathway. Differentially expressed proteins that were simultaneously associated with autophagy, apoptosis, and the HIF1A signaling pathway included BNIP3, HMOX1, SLC2A1, and HK2, all of which are downstream molecules of HIF1A. Protein interaction analysis further demonstrated that BNIP3, HMOX1, and SLC2A1 interact with HIF1A and ATG7.
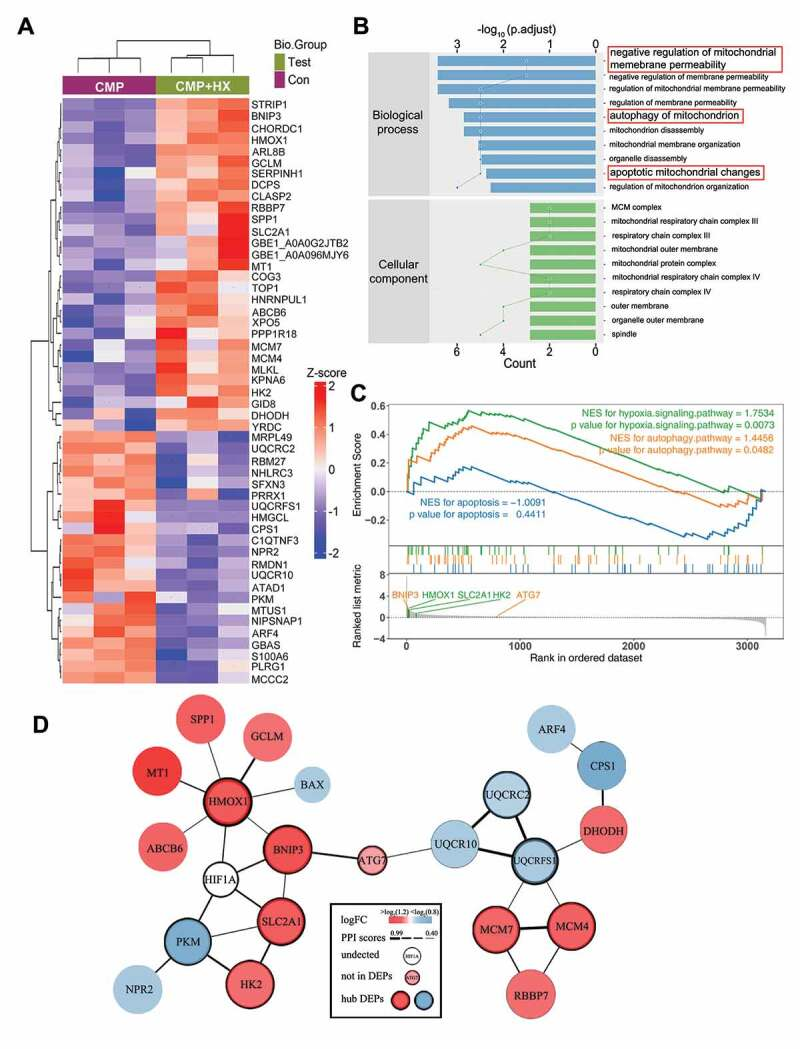
He, R J. et al. Autophagy, 2021.
Figure 2. Quantitative Proteomics Analysis of Proteins Mediating HIF1A Autophagy Stimulation.
MtoZ Biolabs provides proteomics, metabolomics and other technical services based on mass spectrometry, and continues to deepen its efforts in the field of mass spectrometry, with complete proteomic detection and bioinformatics data analysis solutions. In the process of continuous development, MtoZ Biolabs' business scope covers proteome, peptide group, metabolic group, transcriptome, microbiome, biological product characterization, single-cell analysis, bioinformatics cloud analysis, and multi-omics integration analysis, etc., and can be used in multiple angles and in all aspects. The direction provides customers with one-stop scientific research services.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Single Cell Pathway Analysis Service
How to order?







