Proteomics Mass Spectrometry Analysis Service
Proteomics mass spectrometry analysis service utilizes the combined technology of high-performance liquid chromatography (HPLC) and high-resolution mass spectrometry (MS) to separate, detect, and identify all proteins in complex samples. Proteomics is the systematic study of all proteins in a biological system, focusing on their structure, function, interactions, and dynamic changes. It aims to uncover the essence of cellular biology, disease mechanisms, and drug actions. With the advantages of high sensitivity, high resolution, and high throughput, proteomics mass spectrometry analysis has become a pivotal tool for protein structure analysis, quantitative analysis, and interaction studies. MtoZ Biolabs provides proteomics mass spectrometry analysis service including protein identification, relative or absolute protein quantification, protein sequencing, and post-translational modification (PTM) analysis.
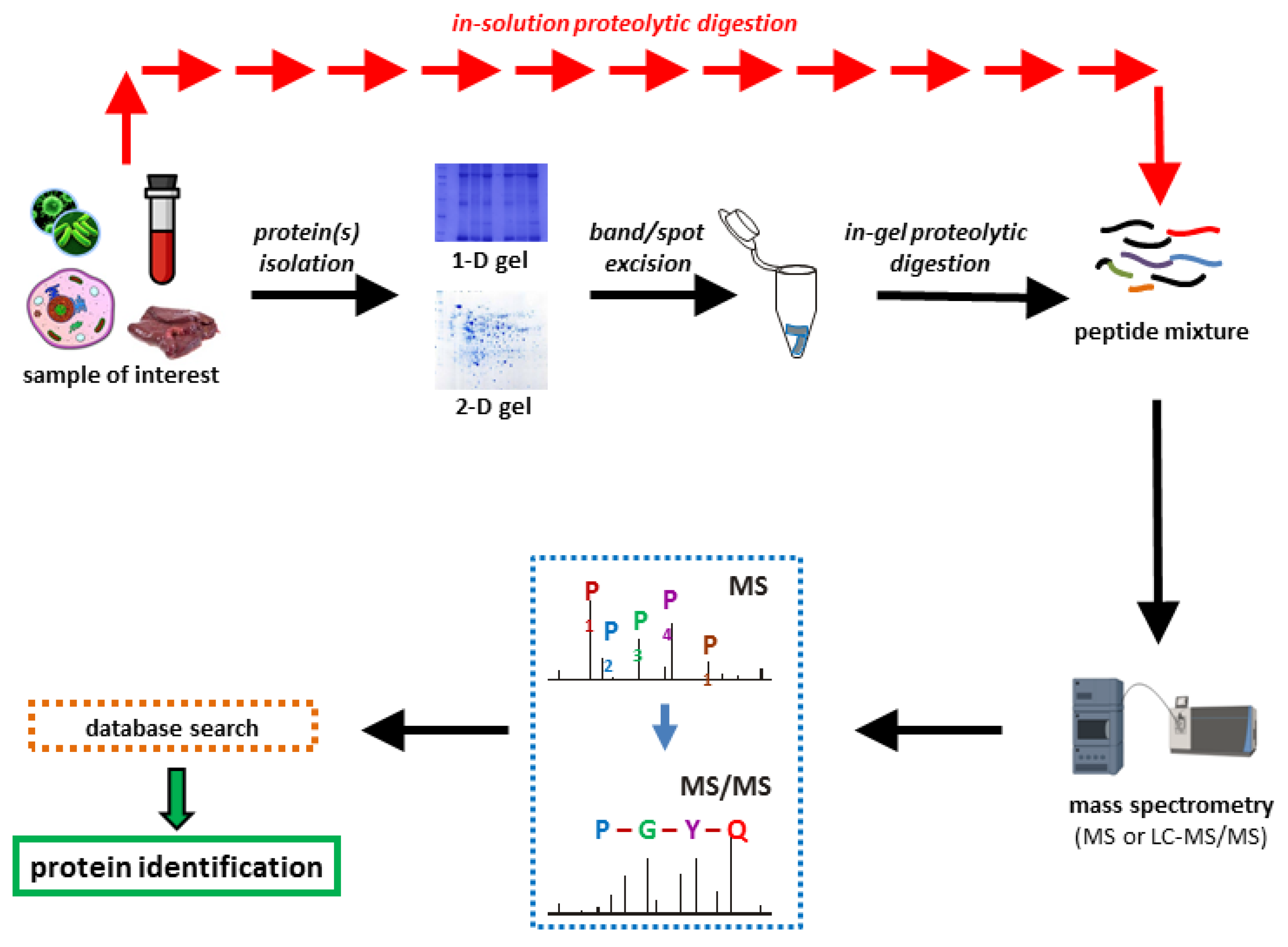
Dupree EJ. et al. Proteomes. 2020.
Analysis Workflow
The analysis workflow for proteomics mass spectrometry analysis generally includes the following main steps:
Sample Preparation: Proteins are extracted from biological samples and digested with enzymes (such as trypsin) into peptides for subsequent analysis.
Peptide Separation: Peptides are separated using techniques such as liquid chromatography to reduce complexity and enhance detection sensitivity.
Mass Spectrometry Analysis: High-resolution mass spectrometer is used to detect the separated peptides, obtaining their mass-to-charge ratio (m/z) data, and further analyzing peptide sequences through fragmentation.
Data Analysis: Bioinformatics tools and databases are used to interpret the mass spectrometry data, identify the proteins corresponding to the peptides, and analyze information about protein quantification, modifications, and interactions.
Results Validation and Analysis: Statistical processing is performed on the analysis results to verify the reliability and accuracy of the data, and to further conduct functional annotation and biological significance exploration.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced proteomics mass spectrometry analysis platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all proteomics mass spectrometry analysis data, providing clients with a comprehensive data report.
Applications
Disease Research and Biomarker Discovery
By analyzing protein expression in different disease states, potential biomarkers can be identified, aiding in early diagnosis, disease monitoring, and the identification of therapeutic targets.
Drug Development
Proteomics mass spectrometry analysis can be used to identify drug targets, study drug mechanisms, and detect drug-target interactions, accelerating the discovery and development of new drugs.
Cell and Molecular Biology Research
By identifying and quantitatively analyzing proteins, changes in protein expression under different physiological states within cells can be studied, exploring intracellular signaling pathways, protein interactions, and metabolic pathways.
Clinical Applications
Proteomics mass spectrometry analysis can be used for disease screening, designing personalized treatment plans, and detecting abnormal proteins in clinical samples (such as in cancer or metabolic diseases).
Food and Environmental Analysis
In fields like food safety testing and environmental pollutant analysis, proteomics mass spectrometry analysis helps identify and quantify protein components and their changes in food, ensuring food safety and quality.
FAQ
Q: How to efficiently extract proteins from various types of samples while preventing sample degradation or contamination?
Ensuring efficient protein extraction and preventing sample degradation hinges on optimizing the sample handling conditions and taking appropriate protective measures.
Choose the appropriate extraction buffer: Select the right buffer based on the type of sample. The buffer should include an appropriate amount of detergents to enhance cell lysis efficiency while avoiding protein degradation.
Protease inhibitors and antioxidants: Add protease inhibitors (such as PMSF, EDTA, or AEBSF) during the extraction process to prevent protein degradation. Additionally, incorporating antioxidants (such as DTT, TCEP, or β-mercaptoethanol) can prevent protein damage caused by redox reactions.
Cold temperature operations: Always perform extraction and processing at low temperatures (e.g., on ice) to minimize the impact of temperature on protein stability.
Timely sample handling and storage: Proteins extracted should be used immediately for analysis or be frozen for storage. For long-term storage, aliquot and store samples at -80°C to avoid degradation or changes due to repeated freeze-thaw.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
In the study of glioblastoma (GBM), TRIM22 protein was found to significantly affect the proliferation of GBM and its response to the chemotherapy drug TMZ (Temozolomide) by regulating the modification and degradation of RIG-I (Retinoic acid-inducible gene I). In order to gain a deeper understanding of the specific role of TRIM22 in this process, the researchers conducted high-throughput protein spectrum analysis on glioblastoma cell lines and TMZ-treated cell samples using proteomics mass spectrometry analysis technology, capturing the key regulatory proteins of TRIM22 in the process of regulating RIG-I, and identifying a series of downstream effector molecules related to the action of TRIM22, revealing the important role of TRIM22 in tumor progression, and also providing new ideas for the development of targeted therapies based on the TRIM22 and RIG-I pathways.
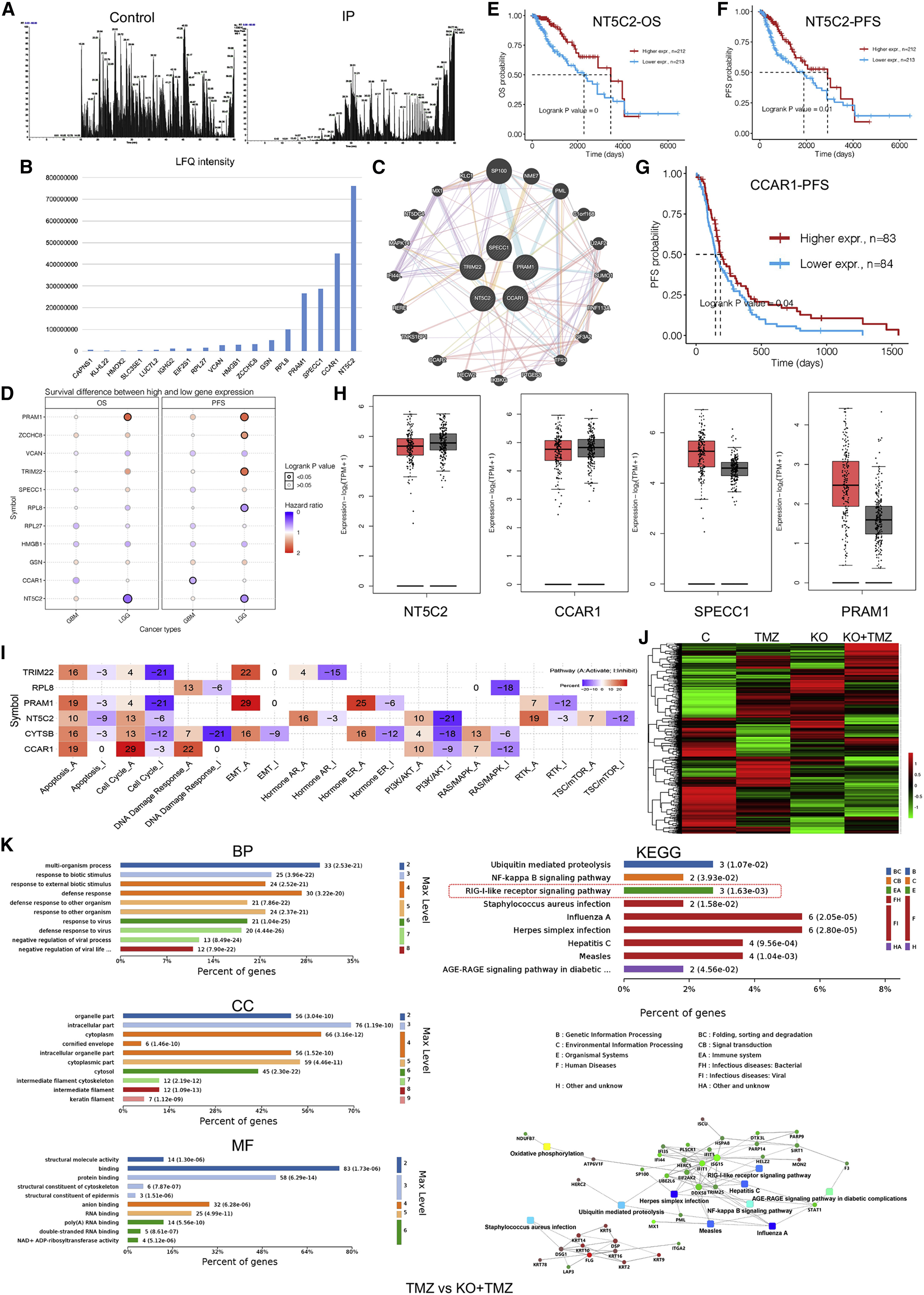
Fei XW. et al. Molecular Therapy Oncolytics. 2022.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Protein Sequencing Service by Mass Spectrometry
Mass Spectrometry-Based Protein Identification Service
Mass Spectrometry-Based Protein Modification Sites Analysis Service
How to order?







