Protein-Protein Interaction Analysis Services
Protein-Protein Interaction Analysis Service is designed to systematically identify and validate the physical binding or functional associations between two or more proteins. Its core objective is to elucidate how proteins interact to form stable or dynamic complexes, thereby regulating a variety of biological processes, including signal transduction, transcriptional regulation, metabolic pathways, structural assembly, and immune responses.
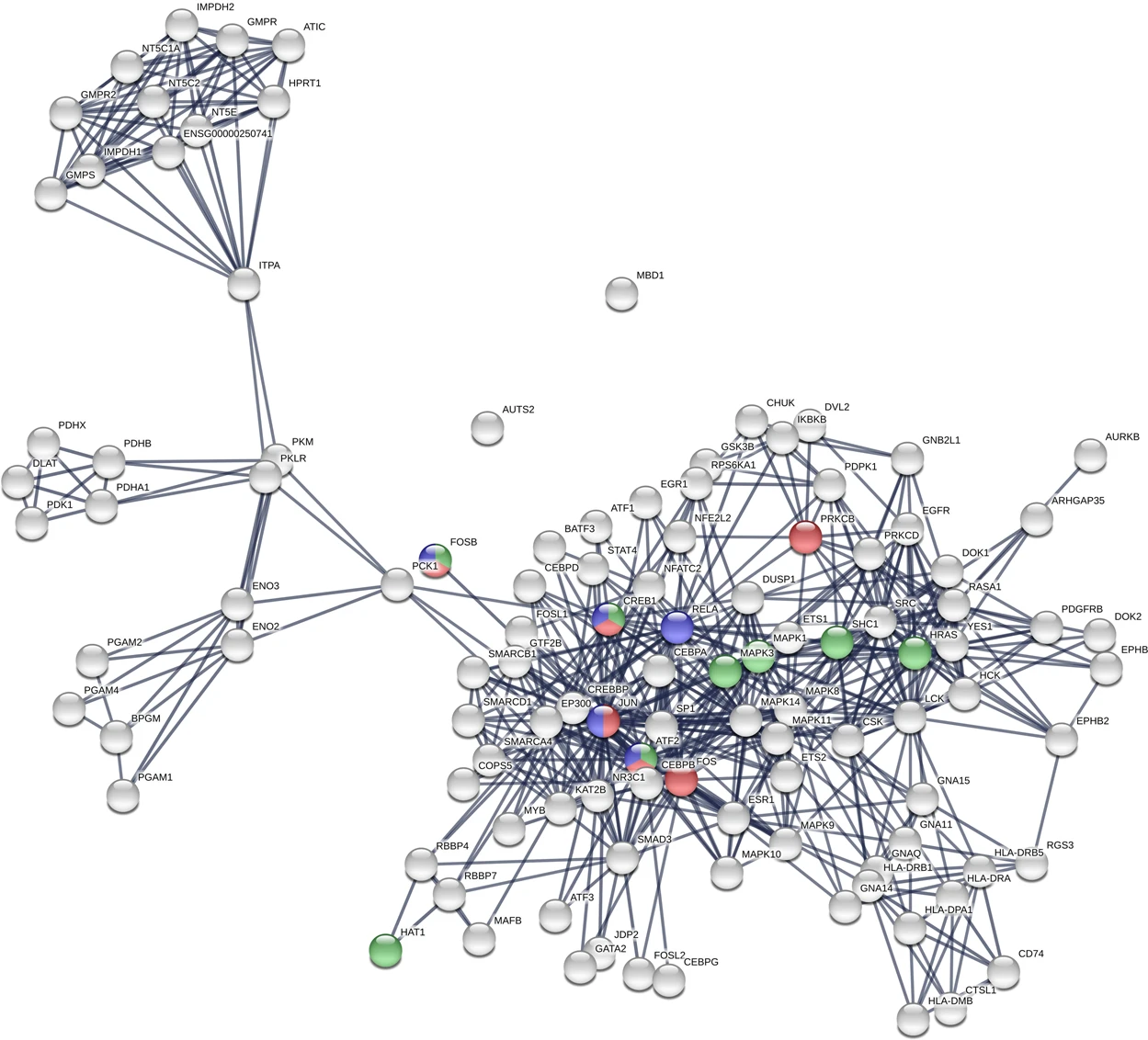
Chen SJ. et al. Sci Rep. 2019.
Protein-protein interactions refer to the physical associations formed between two or more protein molecules through non-covalent bonds, such as hydrogen bonds, hydrophobic interactions, and electrostatic forces. These interactions can be direct (e.g., dimers or complexes) or indirect (e.g., mediated by scaffold proteins to form functional networks). Within cells, proteins interact to form stable or dynamic networks that carry out a wide range of biological functions, including signal transduction, metabolic regulation, cell cycle control, gene expression, and immune responses. Abnormalities in protein-protein interactions (PPIs) are closely associated with various diseases, such as cancer, neurodegenerative disorders, and immune system dysfunctions. Therefore, systematic investigation of PPIs is crucial for understanding fundamental biological processes, uncovering disease mechanisms, and developing targeted therapeutics.
MtoZ Biolabs offers Protein-Protein Interaction Analysis Services that integrates both screening-based and validation-based strategies. This service enables qualitative identification, quantitative measurement, and interaction network construction of protein partners, helping clients systematically dissect protein interaction networks. From basic research to translational applications, we provide full support for scientific discovery and drug development.
Technical Principles
Protein-Protein Interaction Analysis Service can be categorized into screening-based and validation-based approaches depending on the strategy employed:
Yeast Two-Hybrid (Y2H) System: By fusing candidate proteins to a transcriptional activation domain and a DNA-binding domain, respectively, and co-expressing them in yeast cells, interaction between the two proteins drives reporter gene expression. This is a classical high-throughput method for screening potential interaction partners.
Co-Immunoprecipitation (Co-IP): This method uses specific antibodies to enrich target protein complexes, followed by identification and quantification of co-precipitated proteins via mass spectrometry or Western blotting. It is widely used for validating interactions under native physiological conditions.
GST Pull-Down Assay: The "bait protein" is expressed as a fusion with glutathione S-transferase (GST), and the "prey protein" is captured on affinity columns. This assay is suitable for investigating direct binding interactions in vitro.
BiFC and Fluorescence Co-localization: These techniques utilize fluorescent signal reconstitution or co-localization patterns to provide spatial validation and subcellular localization evidence of protein interactions.
Each of these methods has its own advantages and can be combined to improve the coverage and reliability of interaction analysis.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Protein-Protein Interaction Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Protein-Protein Interaction Analysis Service data, providing clients with a comprehensive data report.
Applications
The Protein-Protein Interaction Analysis Service can be applied in the following areas:
Disease Mechanism Research
Investigating the interaction networks of pathogenic gene products to uncover aberrant signaling pathways.
Drug Target Discovery and Validation
Identifying protein complexes or downstream regulatory factors involved in drug action to support target screening and validation.
Signal Pathway Analysis
Reconstructing key signaling networks and studying how post-translational modifications influence protein interactions.
Systems Biology
Integrating multi-omics data to build tissue-specific or disease-specific protein interaction maps.
Synthetic Biology and Protein Engineering
Validating the interaction functions of artificially designed proteins and evaluating their structural stability.
FAQ
Q. Can Protein-Protein Interactions under Specific Post-Translational Modification (PTM) States, such as Phosphorylation-Dependent Binding, be Identified?
Yes. We offer an integrated solution that combines PTM enrichment with interaction analysis. For example, in Co-IP or pull-down experiments, phosphorylated peptides can be enriched using TiO₂ or IMAC prior to interaction profiling, allowing identification of modification-dependent binding events. We also support comparative interaction studies using site-directed PTM mutants to reveal how specific modifications regulate interaction networks. This approach is especially valuable in studying kinase pathways, signal transduction, and drug mechanisms, and is widely applied in cancer and immunology research.
Q. Is it Possible to Perform Protein-Protein Interaction Analysis with Limited Sample Amounts?
Yes. MtoZ Biolabs has well-established protocols for low-input samples, including tag-based enrichment strategies coupled with high-sensitivity mass spectrometry. For rare cell lines, primary cells, or clinical samples, we optimize lysis and immunoprecipitation conditions and minimize non-specific background to ensure reliable identification of interacting proteins.
Case Study
This study systematically investigated how pathogenic mutations at human phosphorylation sites affect protein-protein interactions (PPIs). Focusing on several disease-associated phosphosite mutations—particularly those in the transcription factor GATAD1—the researchers found that its interaction with the 14-3-3 protein family depends on phosphorylation at a specific site. Mutations at this site disrupt the phosphorylation modification, thereby significantly weakening the interaction with 14-3-3 proteins. These findings indicate that pathogenic mutations can interfere with phosphorylation-mediated PPI networks, representing a key molecular mechanism underlying disease development.
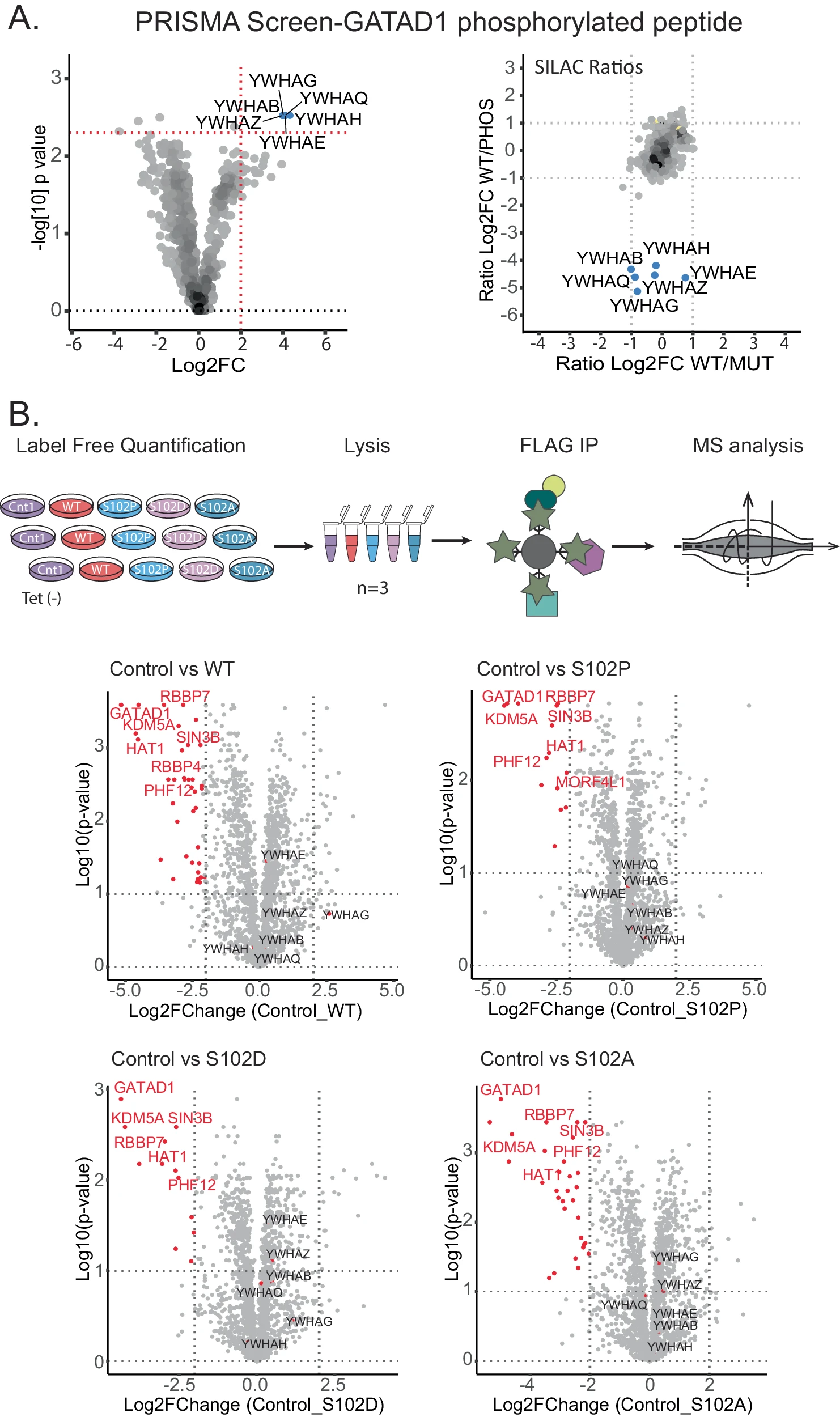
Rrustemi T. et al. Nat Commun. 2024.
How to order?







