Protein PEGylation Analysis Service
Protein PEGylation analysis is a powerful approach for systematically characterizing the structural features, modification sites, and heterogeneity of PEG-modified proteins, as well as their impact on biological function. By integrating quantitative mass spectrometry and structural assessment techniques, researchers can gain in-depth insights into how PEGylation influences protein conformation, stability, pharmacokinetics, and immunogenicity—key factors for therapeutic protein design and quality control.
PEGylation, a widely applied bioconjugation strategy, enhances protein solubility, thermal stability, and circulation half-life. As a modification that profoundly alters protein physicochemical properties and higher-order structures, PEGylation has become central to the development of long-acting therapeutics, sustained-release formulations, and low-immunogenic delivery platforms.
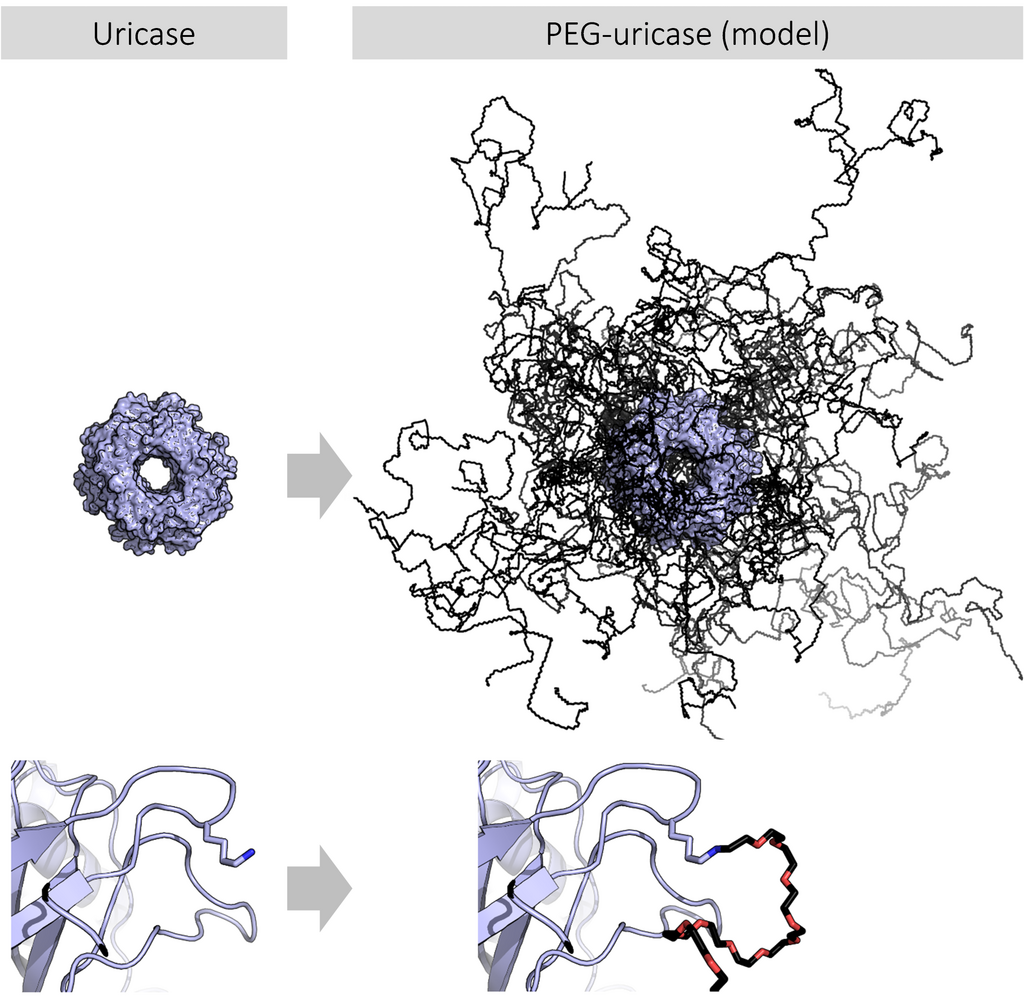
Source: Wikipedia
Figure 1. Structural Comparison of Native and PEGylated Uricase (Model)
MtoZ Biolabs offers a fully quantitative, end-to-end Protein PEGylation Analysis Service by integrating high-resolution mass spectrometry, liquid chromatography, and NMR-based structural analysis. Our platform enables comprehensive site-specific characterization, structural integrity evaluation, and heterogeneity profiling, providing a one-stop solution for PEGylated protein analysis in both research and manufacturing contexts.
Services at MtoZ Biolabs
1. Targeted Protein PEGylation Analysis
Designed for studies focusing on specific proteins, this service identifies and quantifies PEG attachment sites on client-provided targets. Following proteolytic digestion and peptide-level enrichment, high-resolution LC-MS/MS is used to confirm PEGylation patterns with high accuracy.
2. PEGylation Proteomics
Intended for global investigations, this service profiles PEGylation modifications across the proteome. Using large-scale LC-MS/MS with peptide enrichment, it supports comparative or discovery-driven research across tissues, cell types, or treatment conditions.
Analytical Approaches
Our core analytical approaches include:
· High-Resolution LC-MS/MS
Using Orbitrap-based systems and tailored enzymatic digestion, we perform precise identification of PEGylation sites, chain length analysis, and quantification. Both bottom-up and top-down strategies are available for comprehensive molecular characterization.
· Size-Exclusion Chromatography (SEC)
Separates PEGylated variants by molecular size to assess heterogeneity, aggregation, and molecular weight distribution. Facilitates purity assessment for downstream MS analysis.
· Reverse-Phase HPLC (RP-HPLC)
Evaluates changes in protein hydrophobicity and retention behavior post-PEGylation, supporting process consistency and conformation monitoring.
· NMR Spectroscopy (Optional)
Provides tertiary structure insights, including domain flexibility, epitope exposure, and potential steric interference caused by PEG conjugation.
· Western Blot/ELISA (Supplementary Validation)
Validates modification sites and functional outcomes through site-specific antibody recognition or post-modification activity assessment.
Why Choose MtoZ Biolabs?
✔ Unmatched Site Resolution: Detect exact PEGylation positions and evaluate proximity to key structural or functional motifs
✔ Quantitative Heterogeneity Deconvolution: Profile PEG chain-length distribution and site combinations across product batches
✔ Cross-Platform Integration: Correlate data from MS, SEC, HPLC, and NMR for complete structural and functional insight
✔ Broad Strategy Compatibility: N-terminal, lysine, or thiol-specific PEGylation; supports biologics, biosimilars, and custom conjugates
✔ High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrate all proteomics analysis data, providing clients with a comprehensive data report.
✔ One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
Applications
Protein PEGylation Analysis Service is widely used in PEG modification related drug development, structure validation and modification mechanism studies, including:
· Formulation optimization and drug delivery: Assess how PEGylation enhances plasma stability and circulation time
· Immunogenicity prediction and risk control: Identify PEGylation sites that may alter epitope accessibility or trigger immune responses
· Long-acting biologics development: Guide site selection and validate conjugation strategies in half-life extension projects
· Lot-to-lot comparability and stability tracking: Perform inter-batch structure profiling and aging-based modification surveillance
· Filing-ready structural documents: Provide PEGylation maps and characterization files for sustained-release drugs, ADC linkers, or biosimilar submissions
Case Study
Case 1: Mass Spectrometry–Based Profiling of PEGylated Proteins in Complex Matrices
A 2024 study established an LC–MS/MS method for detecting and quantifying PEG-conjugated moieties in plasma and tissue samples. Acid hydrolysis was used to release PEG signature fragments, which were then sensitively detected by targeted MS workflows.
This method proved effective across PEG-modified nanoparticles, lipid-based therapeutics, and PEGylated proteins—demonstrating utility in vivo pharmacokinetics, drug clearance profiling, and QC validation. The approach was validated for both human and preclinical samples.
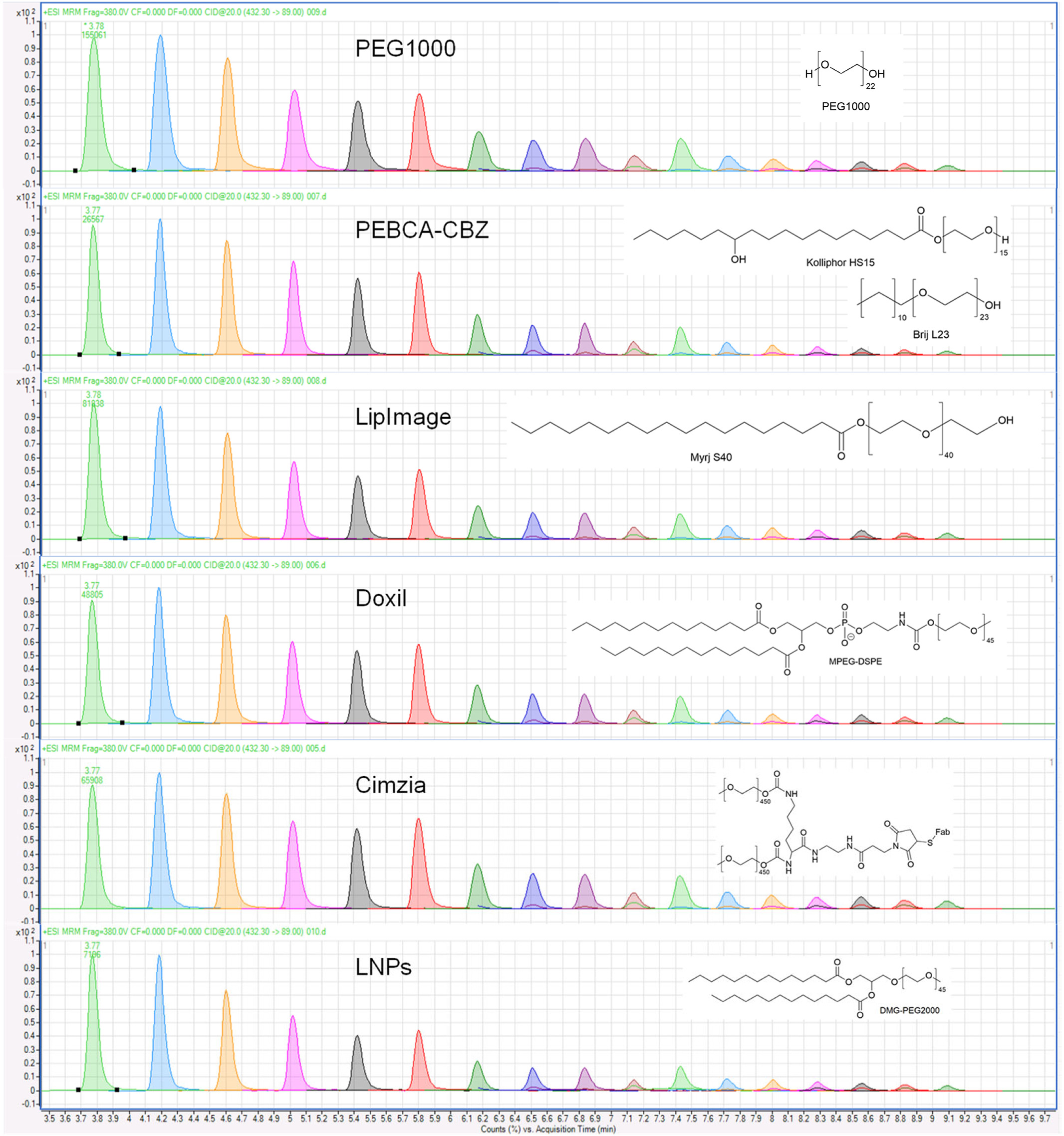
Hyldbakk, A. et al. J. Control. Release. 2024.
MtoZ Biolabs' Protein PEGylation Analysis Service provides reliable modification assessment and in vivo behavioral characterization of PEG-modified proteins by leveraging a high-precision mass spectrometry platform with optimized sample processing.
FAQ
Q1: How do you distinguish between N-terminal, lysine, and cysteine PEGylation? Are there analytical limitations?
Yes, each conjugation site has distinct reactivity and steric environment, requiring tailored digestion and enrichment strategies. We apply site-targeted enzymatic workflows and tandem MS to confidently assign PEG types. Our reports include detailed annotations of linkage chemistry and position, aiding structure-function modeling and optimization.
Q2: Can PEGylation interfere with protein function? How can this be evaluated?
Indeed. PEG chains can block receptor-binding interfaces, catalytic pockets, or antibody epitopes. We offer structural overlay analysis using NMR or AlphaFold models to evaluate steric interference. These data help refine conjugation strategies for minimal functional disruption.
Q3: Do you support low-input sample analysis?
Yes. With nanoLC-MS and optimized handling, we can analyze as little as 50–100 µg of protein while still resolving PEGylation patterns and structural variants—ideal for rare, clinical, or early-stage development samples.
MtoZ Biolabs’ Protein PEGylation Analysis Service provides a full-spectrum solution for characterizing PEG-modified proteins—combining structural precision with quantitative depth to support functional discovery and regulatory alignment. Contact us for tailored consultation and project design.
How to order?







