Protein Backbone PTM Analysis Service
Protein Backbone PTM Analysis Service refers to a service that utilizes high-resolution mass spectrometry and related technologies to systematically identify, characterize, and quantify post-translational modifications (PTMs) occurring on the protein backbone, in order to elucidate their regulatory mechanisms on protein conformation, functional states, and molecular recognition.
The structure and function of proteins are determined not only by their amino acid sequences but also by PTMs. While most studies have focused on modifications of side-chain residues—such as lysine, serine, threonine, and tyrosine—recent research has revealed that PTMs occurring on the protein backbone also possess significant biological relevance. Backbone modifications refer to chemical groups (e.g., hydroxyl, amide, imine) that directly target the peptide bond, nitrogen atom, or carbonyl oxygen of the backbone, thereby affecting peptide chain conformation, secondary structure formation, stability, and protein–protein interactions. These modifications include N-terminal backbone modifications (e.g., N-formylation, N-methylation), C-terminal carboxyl backbone modifications (e.g., esterification, hydroxylation), and noncanonical amide bond formation (e.g., cyclization). They have been implicated in biological processes such as bacterial virulence, programmed cell death, and signal transduction.
Müller MM. Biochemistry. 2018.
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs provides the Protein Backbone PTM Analysis Service focusing on the identification of PTM occurring on the protein backbone. This service enables precise localization and quantitative analysis of modification sites, supporting researchers in exploring the mechanisms of conformational regulation, structural stability, and functional diversity of proteins.
Analysis Workflow
The analytical workflow of Protein Backbone PTM Analysis Service is as follows:
1. Sample Preparation
Samples are pretreated and digested with proteases according to sample type.
2. Modification Enrichment/Labeling
Specific chemical labeling reagents or affinity capture methods are selected based on the target modification to enhance the proportion of backbone-modified peptides.
3. LC-MS/MS Analysis
Peptides are separated and fragmented using a high-resolution mass spectrometry platform to obtain MS1 and MS2 spectra.
4. Data Search and Modification Identification
Open modification search strategies are applied to identify backbone PTM sites. Manual spectrum validation and peptide verification ensure identification accuracy.
5. Quantitative Analysis and Functional Annotation
Relative or absolute quantification is performed for modification sites, followed by biological interpretation based on protein domains, conservation, and protein–protein interaction (PPI) networks.
Applications
Protein Backbone PTM Analysis Service can be applied in a wide range of research areas, including:
Structural Elucidation of Novel Natural Products
Characterize cyclic peptides, amide bridges, and noncanonical acyl structures to support structural validation and optimization of bioactive peptide drugs.
Pathogenicity Factor Studies in Bacteria or Viruses
Identify backbone modifications in pathogenic proteins to investigate their roles in host recognition or infectivity.
Stability Evaluation and Modification Design of Peptide Drugs
Monitor modifications such as backbone hydroxylation and amide isomerization that affect stability, providing guidance for modified drug development.
Studies on Protein Conformation and Secondary Structure Regulation
Reveal how backbone modifications influence α-helix and β-sheet formation to advance integrative structure–function analysis.
FAQ
Q. Can Backbone Modifications be Distinguished from Side-Chain Modifications, Especially When Occurring at Neighboring Residues?
Yes. Using high-resolution mass spectrometry combined with b/y ion series fragmentation, MtoZ Biolabs can accurately localize modification sites. Open search strategies and manual spectrum interpretation further enable reliable differentiation between backbone and side-chain modifications at adjacent residues, minimizing misannotations and ensuring confident site identification.
Q. Are there Specific Chemical Enrichment or Derivatization Strategies for Noncanonical Backbone Modifications such as Amide Isomerization or Hydroxylation?
Yes. For amide and hydroxyl group modifications on the backbone, we employ targeted derivatization reagents—such as difluoroamides and isocyanate-based probes—for chemical labeling and enrichment, enhancing the MS detectability of these modifications. These strategies significantly improve the identification of low-abundance backbone modifications that are typically difficult to capture using conventional enrichment methods, particularly in natural peptides or cyclic structures.
Case Study
This study systematically analyzed a backbone isomerization modification in the essential bacterial enzyme MurA, specifically the formation of β-isoaspartate (isoAsp) at Asn67. Unlike conventional interpretations of isoAsp as a marker of protein aging, this modification was found to form spontaneously at an unusually rapid rate in MurA and positively contribute to the enzyme’s folding stability. Mutation or inhibition of the isomerization process led to protein aggregation and loss of function, highlighting the critical role of isoAsp in maintaining MurA’s three-dimensional structure and biological activity.
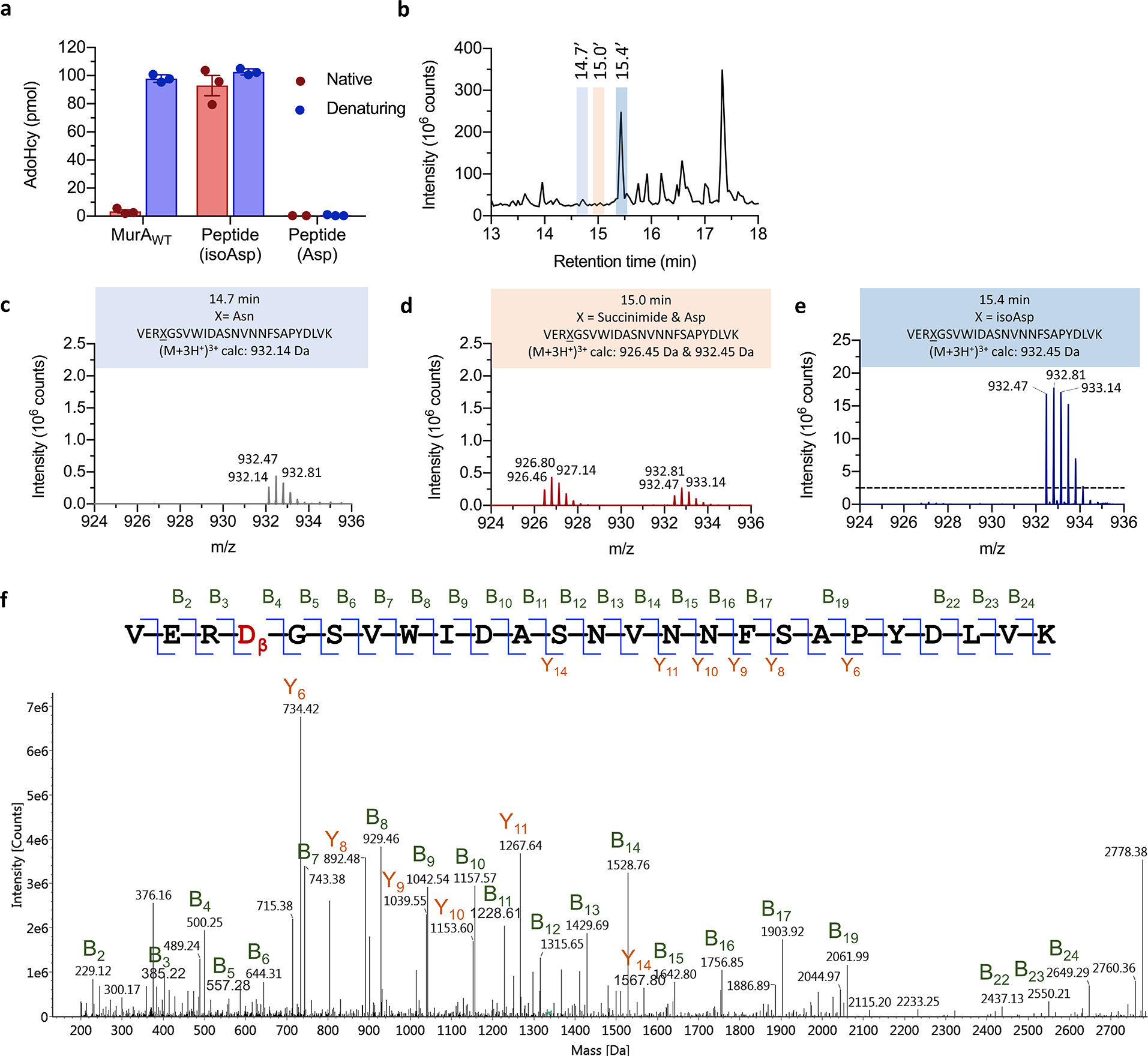
Zhang T. et al. Biochemistry. 2020.
How to order?







