Protein and Protein Interaction Analysis Service
Protein and protein interaction analysis refers to the process of studying the interactions between different proteins and their functional relationships using experimental techniques and methods. Protein-protein interactions are central to biological processes within cells, including signal transduction, metabolism, gene regulation, cell division, and immune responses. The core aspects of protein and protein interaction analysis service include identifying interacting proteins, investigating interaction mechanisms, performing functional analyses, and constructing interaction networks. Leveraging advanced chromatography and mass spectrometry technologies, MtoZ Biolabs offers protein and protein interaction analysis service aiming to identify and characterize interacting proteins to lay a solid foundation for further studies on interaction mechanisms, functional analyses, and interaction network construction.
Analysis Workflow
General workflow for protein and protein interaction analysis service:
1. Sample Preparation
Collect and process target proteins, followed by appropriate purification and labeling.
2. Interaction Screening
Screen target proteins and their potential interacting partners using techniques such as immunoprecipitation or co-immunoprecipitation.
3. Mass Spectrometry Analysis
Identify and quantify the screened proteins using high-resolution mass spectrometry technology.
4. Data Analysis
Analyze protein interaction networks using bioinformatics tools.
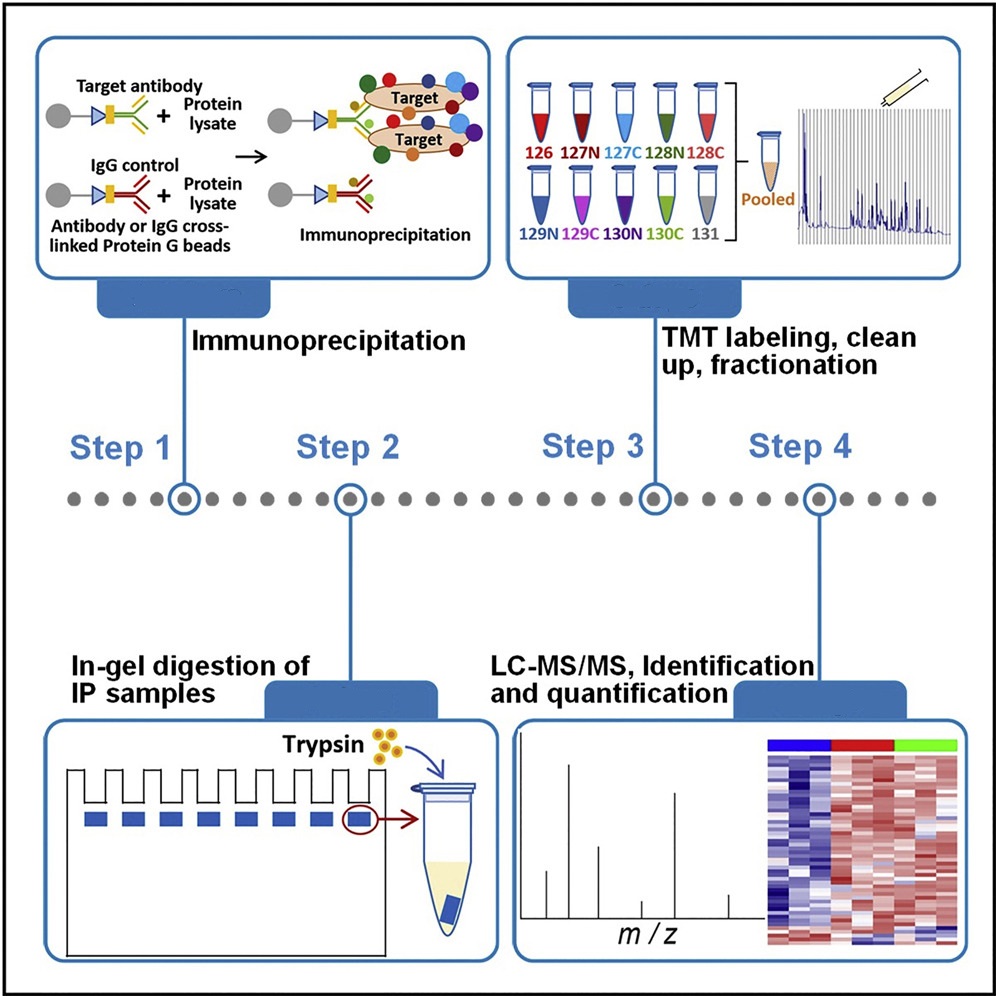
Kumar R. et al. STAR protoc. 2022.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs provides protein interaction analysis and subsequent LC-MS/MS-based analysis for protein/protein mixtures in purified samples such as IP, Co-IP and GST fusion protein pull-down samples.
Applications
Applications of protein and protein interaction analysis service:
Cell Signaling
Identify key regulatory proteins and their interaction networks to explore the mechanisms of signal transduction pathways.
Protein Complex Analysis
Reveal the assembly and functions of multiprotein complexes, uncovering their roles in cellular processes.
Disease Research
Utilize protein interaction maps to identify interactions involving pathological proteins, providing insights into potential therapeutic targets.
FAQ
Q. How to Distinguish Specific from Non-Specific Protein Interactions?
Distinguishing specific protein interactions from non-specific ones is crucial for ensuring the accuracy of experimental data. The following methods can effectively address this challenge:
1. Set Appropriate Controls
Negative Control: Use unrelated antibodies (e.g., IgG) in the experiment to exclude non-specific binding.
Non-Target Protein Control: In co-immunoprecipitation experiments, use samples that do not express the target protein as a control.
2. Optimize Experimental Conditions
Adjust Buffer Composition: Include high-salt buffers (e.g., 300–500 mM NaCl) or non-ionic detergents (e.g., Triton X-100) in washing steps to remove weak non-specific background proteins.
Competition Experiments: Validate interaction specificity by introducing free target proteins or antibodies to compete with binding.
3. Repeat Experiments and Multi-Technique Validation
Repeat Experiments: Conduct interaction analyses under different experimental conditions to ensure consistent and specific results.
Multi-Technique Validation: Use complementary methods such as FRET (Förster Resonance Energy Transfer), SPR (Surface Plasmon Resonance), or Bimolecular Fluorescence Complementation (BiFC) to confirm specific protein interactions.
4. Employ Quantitative Mass Spectrometry
Labeled Quantification (e.g., TMT or iTRAQ): Quantitatively compare target protein interactions with control groups to identify high-confidence specific interactions.
Label-Free Quantification (LFQ): Combine high-resolution mass spectrometry with data analysis software to further validate interaction protein reliability.
5. Utilize Bioinformatics-Assisted Analysis
Leverage protein databases (e.g., STRING, BioGRID) for functional annotation and network analysis to confirm whether the interaction has known biological relevance.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
The study used mass spectrometry and co-immunoprecipitation techniques for protein-protein interaction analysis to systematically analyze the protein interaction network of Huntington protein (HTT) in normal and disease states. The results showed that HTT's interaction partners changed significantly during the disease process, and these dynamic changes were closely related to cell signaling, protein homeostasis, and neural function regulation.
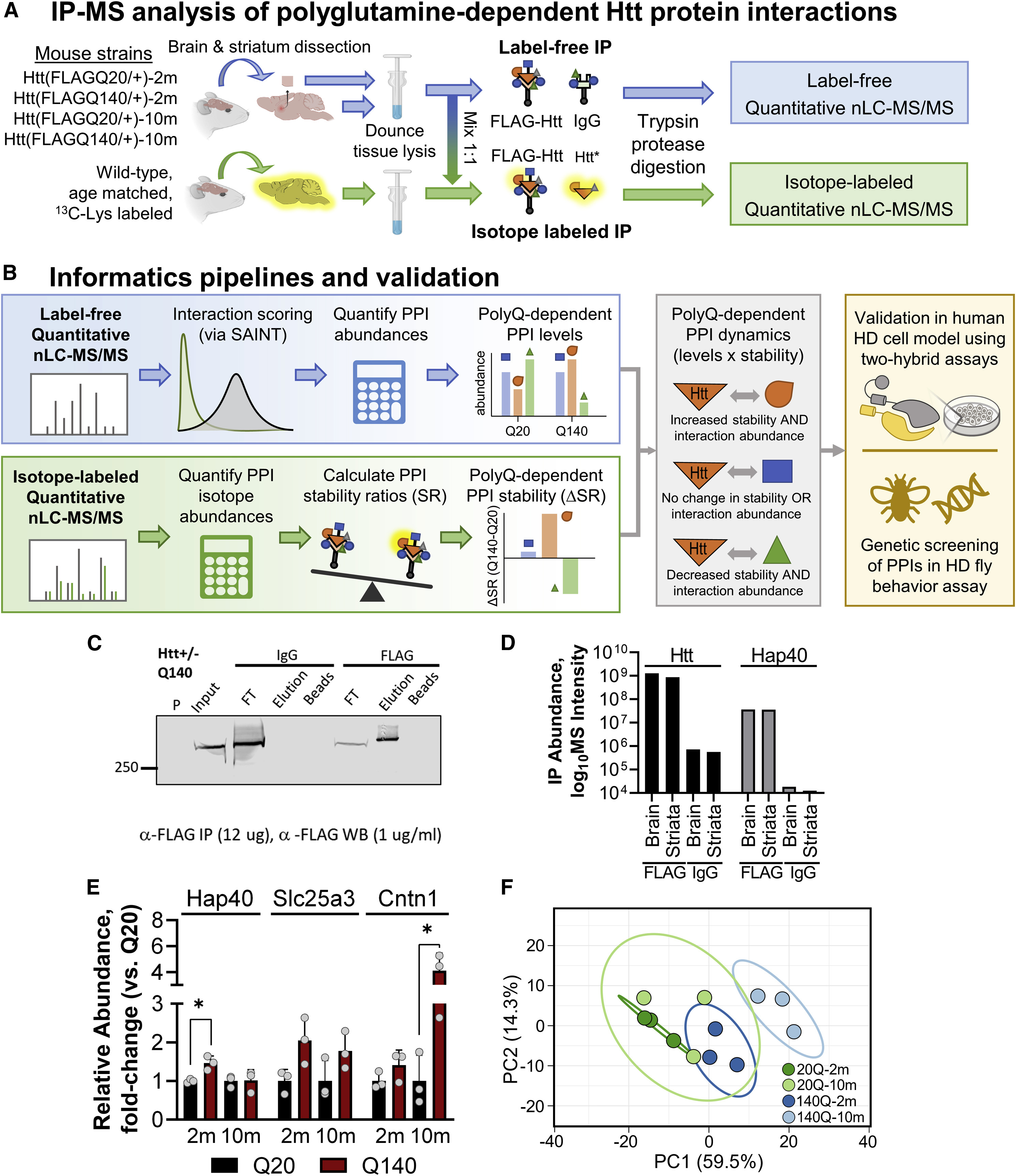
Greco T M. et al. Cell systems. 2022.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
MS-Based Protein-Protein Interaction Analysis Service
How to order?







