Post-Translational Modification Proteomics Solutions
- Cancer: Aberrant phosphorylation of kinases such as EGFR and HER2 drives tumor proliferation.
- Neurodegenerative Diseases: Excessive phosphorylation of Tau protein leads to neurofibrillary tangles in Alzheimer’s disease.
- Metabolic Syndrome: Mitochondrial protein acetylation imbalance is closely associated with insulin resistance.
- Phosphorylation: TiO₂ microspheres selectively bind phosphate groups under acidic conditions.
- Acetylation: Immunoaffinity columns enrich acetylated lysine sites, followed by LC-MS/MS quantification.
- Glycosylation: Hydrophilic interaction chromatography (HILIC) separates glycan variants for detailed characterization.
- Instrument Configuration: Thermo Fisher Orbitrap Exploris™ 480 (Resolution: 480,000 @ m/z 200)
- Scanning Mode: Data-Dependent Acquisition (DDA) and Data-Independent Acquisition (DIA) in parallel, ensuring low-abundance signal detection.
- Mass Accuracy: <2 ppm (peptide level), <0.02 Da (fragment ions)
- Database Search: UniProtKB/Swiss-Prot database + PTM-specific modification libraries
- Quantitative Analysis: Processed using MaxQuant, with differential site selection criteria (Fold Change ≥1.5, p < 0.05)
- Pathway Modeling: Cytoscape visualization tool used to map PTM-kinase-substrate regulatory networks
The post-translational modification proteomics solutions integrate high-throughput mass spectrometry technology with multidimensional bioinformatics analysis to systematically profile dynamic protein modifications and their spatiotemporal-specific regulatory networks. This approach uncovers the molecular interaction patterns and functional effects of PTMs in cellular signaling pathway reprogramming, epigenetic regulation, and disease pathology mechanisms. Post-translational modifications (PTMs) are one of the core mechanisms of dynamic regulation in living organisms. With over 400 known modification types—ranging from phosphorylation and acetylation to ubiquitination—PTMs play crucial roles in cellular signaling, metabolic regulation, and disease progression.
Studies indicate that more than 80% of cancer-related pathways are directly linked to PTM dysregulation. Furthermore, PTMs have been identified as a key breakthrough point for discovering novel therapeutic targets in neurodegenerative diseases and immune system disorders.
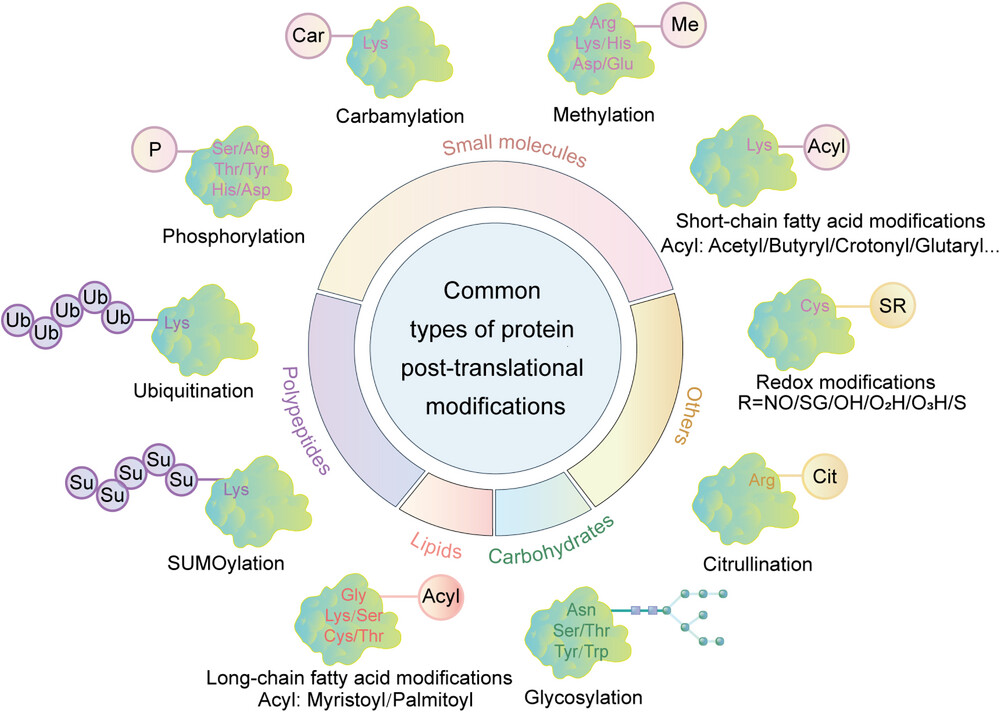
Zhong, Q. et al. MedComm. 2023.
Figure1. Common Types of Protein Post-Translational Modifications
However, PTM research faces multiple technical challenges, including the highly dynamic nature of modification sites, extremely low abundance (with some modified proteins present at concentrations as low as the amol level), and the limited throughput of traditional antibody-based detection methods. Achieving high-sensitivity and high-coverage PTM analysis has become a key priority in global scientific research and drug development.
Service at MtoZ Biolabs
As a specialized service provider in the field of proteomics, MtoZ Biolabs provides end-to-end post-translational modification proteomics solutions through advanced mass spectrometry platforms and proprietary enrichment technologies for global research institutions and biopharmaceutical partners. Our services encompass the entire workflow from PTM discovery to functional validation and clinical translation.
Three Core Services:
1. PTM Discovery Services: Comprehensive profiling of over 20 types of modifications, including phosphorylation, acetylation, and ubiquitination.
2. Targeted Validation Services: Quantitative analysis of key modification sites using PRM/SRM technology.
3. Multi-Omics Integration Services: Construction of regulatory networks by integrating genomics and transcriptomics data.
Service Phases and Key Deliverables:
| Service Phase | Key Deliverables |
|---|---|
| Experimental Design Consultation | Sample preparation strategies, quality control standards, and statistical recommendations |
| Wet-Lab Workflow | PTM enrichment, enzymatic digestion, labeling, and mass spectrometry analysis |
| Data Analysis & Validation | Site identification, functional annotation, and pathway network modeling |
| Clinical Correlation Analysis* | Biomarker screening and drug target prediction |
(*Note: Clinical correlation analysis requires integration with transcriptomics and metabolomics data.)
Analysis Workflow
Sample Preparation → PTM Enrichment → Peptide Digestion → LC-MS/MS → Database Matching → Functional Annotation → Report Generation
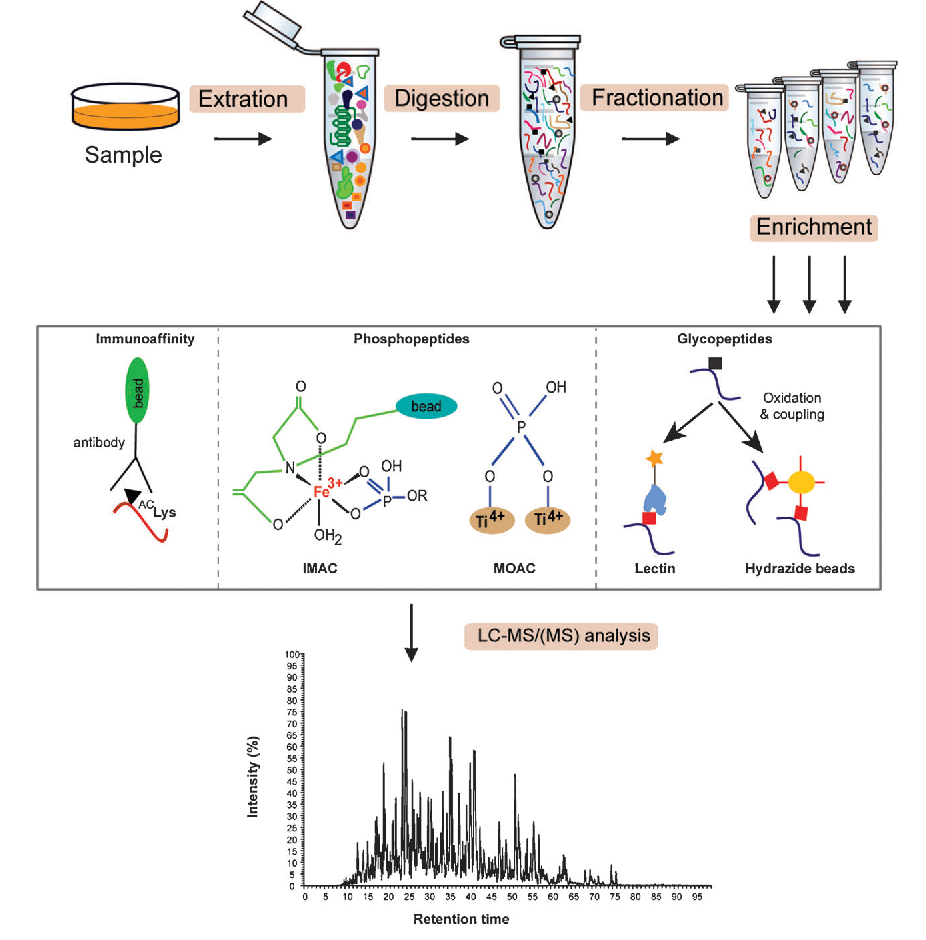
Angel, T. et al. Chem. Soc. Rev. 2012.
Figure2. Experimental Strategies to Study Protein Post-translational Modification
Step 1: Modification-Specific Enrichment
A physicochemical coupling strategy is employed to enhance the capture efficiency of target modifications:
Step 2: Mass Spectrometry Data Acquisition
Step 3: Bioinformatics Analysis
By integrating high-resolution mass spectrometry, advanced bioinformatics, and specialized enrichment strategies, MtoZ Biolabs delivers comprehensive PTM analysis with high sensitivity, precision, and biological relevance.
Applications
✅ Tumor Heterogeneity Analysis: Utilizing spatial PTM proteomics to pinpoint key phosphorylation regulatory nodes in the PD-1/PD-L1 pathway within the tumor microenvironment.
✅ Neurodegenerative Disease Mechanism Research: Developing a Tau protein phosphorylation site database using post-translational modification proteomics solutions to support early biomarker discovery for Alzheimer’s disease.
✅ Drug Toxicity Assessment: Monitoring mitochondrial protein acetylation dynamics under drug intervention to predict cardiotoxicity risks.
Case Study
Post-translational modifications play a crucial role in regulating protein function, structure, and interactions. The heart, a high-energy-demanding organ, involves multiple physiological functions, yet the characteristics of its PTM landscape and disease-associated modifications remain underexplored.
This study employed a post-translational modification proteomics solutions to analyze human cardiac samples using LC-MS/MS. By integrating various separation techniques, affinity enrichment strategies, and chemical labeling methods, researchers identified a wide range of PTMs in cardiac tissue. The study systematically mapped modification sites, modification types, and their distribution across different cardiac tissues and disease states, covering phosphorylation, acetylation, methylation, ubiquitination, and more.
Findings from this study revealed heart-specific PTMs and their alterations in cardiovascular diseases such as myocardial infarction and heart failure, providing critical insights for biomarker discovery and therapeutic target identification.
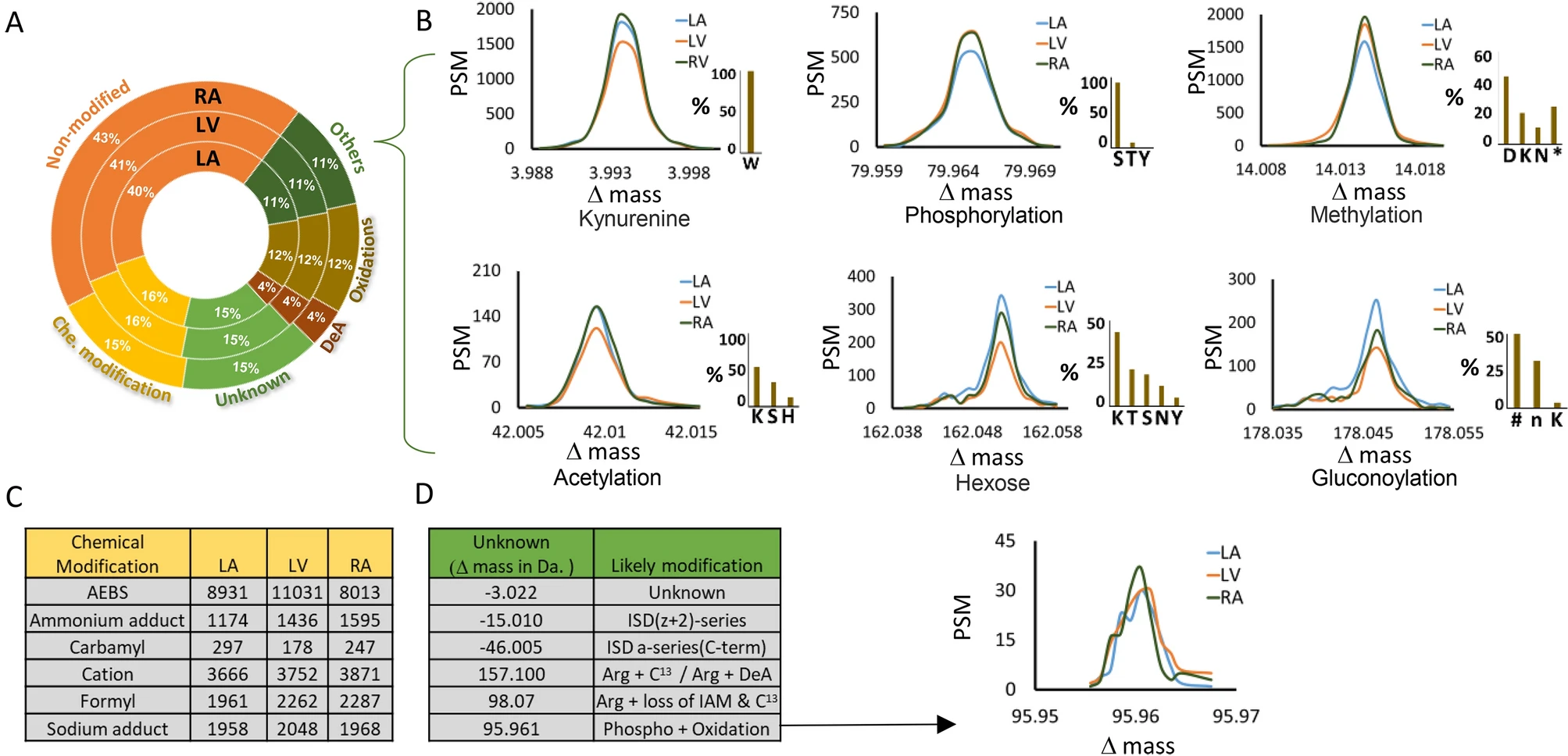
Bagwan, N. et al. Sci Rep. 2021.
Figure3. Classification of Peptide Modifications Across Three Cardiac Chambers
FAQ
Q1: How much sample is required for PTM proteomics analysis?
A: For standard projects, we recommend biological replicates of ≥3, with a minimum of 100 μg of tissue or 1 mL of serum per sample. For precious samples, we offer a micro-sample processing technique, requiring as little as 10 μg.
Q2: Can PTM levels be compared between different treatment groups?
A: Yes, we support TMT/iTRAQ-labeled quantification as well as label-free analysis, capable of detecting fold changes of ≥1.5 with p < 0.05.
Q3: How can PTM data be integrated with genomic variations?
A: We offer multi-omics database integration, including TCGA/ICGC, to map PTM regulatory networks associated with driver mutations.
MtoZ Biolabs is a specialized technology service platform dedicated to post-translational modification proteomics. Our team is led by scientists with publications in Cell, Nature, and other high-impact journals, committed to advancing cancer research, drug development, and precision medicine through innovative PTM analysis solutions.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Other Post-Translational Modification (PTM) Proteomics Service
Global Post-Translational Modification Proteomics Service
Histone Post-Translational Modification (PTM) Proteomics Service
How to order?







