Post-Translational Modification Analysis for Protein Biopharmaceuticals
Post-Translational Modification Analysis for Protein Biopharmaceuticals is a comprehensive analytical service designed to identify, localize, quantify, and functionally annotate various post-translational modifications (PTMs) occurring in protein-based therapeutics. This service covers modification detection at the intact protein, peptide, and even site-specific levels, and is applicable to biologic drug development, process optimization, and quality research.
Protein biopharmaceuticals—such as monoclonal antibodies, fusion proteins, and enzyme preparations—often undergo various PTMs during expression, processing, purification, and storage. These include glycosylation, phosphorylation, deamidation, hydroxylation, acetylation, carboxylation, and more. Such modifications have significant impacts on the structural integrity, stability, biological activity, immunogenicity, and pharmacokinetic behavior of the therapeutic proteins. Variations in PTMs across different production batches or under varying process conditions may lead to product quality deviations, trigger immunogenic responses, or reduce therapeutic efficacy. As a result, PTMs analysis has become a critical quality attribute (CQA) in the development and manufacturing of protein therapeutics, and is a necessary component for regulatory compliance with agencies. Systematic PTMs profiling enables companies to characterize product structural integrity, monitor process consistency, optimize development strategies, and provide essential data for regulatory submissions.
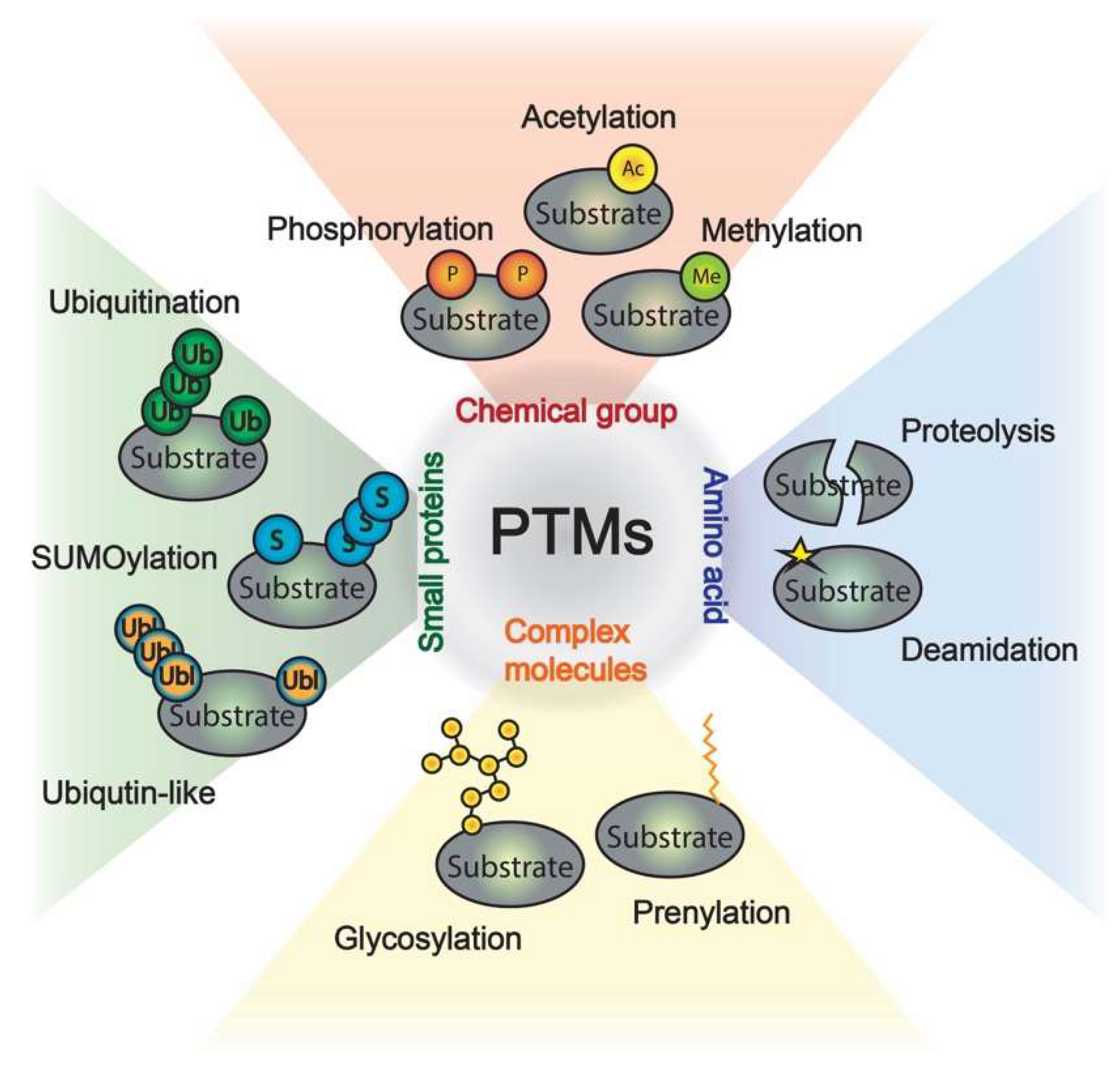
Salas-Lloret D. et al. Int J Mol Sci. 2022.
Leveraging advanced mass spectrometry and chromatography platforms, MtoZ Biolabs provides Post-Translational Modification Analysis for Protein Biopharmaceuticals with high sensitivity for the detection and quantification of diverse PTMs in proteins. This service helps clients gain in-depth insights into modification patterns, ensure product consistency and quality, meet regulatory requirements, and support new drug development and process optimization.
Analysis Workflow
The main analysis workflow for PTMs in protein biopharmaceuticals is as follows:
1. Sample Preparation
Protein samples undergo reduction, alkylation, and enzymatic digestion to generate peptides suitable for mass spectrometry analysis.
2. Enrichment and Separation (Optional)
Based on the modification type, enrichment strategies such as lectin affinity (for glycosylation), TiO₂ chromatography (for phosphorylation), or HILIC (for polar peptides) can be applied to isolate modified peptides.
3. Mass Spectrometry Analysis
High-resolution LC-MS/MS is used to separate and detect peptides, generating high-quality spectral data.
4. Data Processing
Specialized software is used for data analysis to identify and localize PTMs sites on proteins, along with quantification.
5. Functional Annotation and Report Delivery
GO/KEGG annotation of modification sites is performed, and a detailed report is generated to support regulatory submissions and quality assessments.
Applications
New Drug Development
Comprehensive PTMs profiling during biopharmaceutical development helps optimize molecular design and enhance drug stability and efficacy.
Quality Control
Monitoring key PTMs ensures product consistency and quality during manufacturing, meeting regulatory compliance requirements.
Process Optimization
Analyzing modification patterns under different production conditions provides insights for process parameter optimization to improve manufacturing efficiency and product quality.
FAQ
Q. Does the Modification Identification have Site Specificity? Can PTMs be Localized to Specific Amino Acid Residues?
MtoZ Biolabs utilizes high-resolution mass spectrometry platforms in combination with multiple fragmentation modes (e.g., HCD and ETD) to ensure both modification type identification and residue-level localization. Database searches are configured with defined mass shifts for each PTM (e.g., +15.995 Da for oxidation, +0.984 Da for deamidation), and modification sites are pinpointed using fragment ion information (e.g., b/y ions, c/z ions). For structural isomers (e.g., Asp/isoAsp), we incorporate retention time analysis, isomeric peak deconvolution, and diagnostic fragment ions to ensure accurate and specific site identification.
Q. Is Quantitative Comparison of PTMs Across Batches or Bioprocess Stages Supported?
Yes, we support multiple quantification strategies, including Label-Free Quantification (LFQ) and isotopic labeling methods (e.g., TMT, iTRAQ), suitable for comparing PTM levels across batches or process conditions. Standardized workflows and internal quality controls—such as QC sample insertion and cross-batch normalization—are employed to ensure high reproducibility and comparability of abundance data. We also offer statistical analysis and significance testing, delivering standardized reports suitable for quality trend monitoring, process development, and regulatory submission.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
How to order?







