mRNA 5' Capping Rate Analysis Service
mRNA vaccines offer numerous benefits, including cost-effectiveness, significant R&D flexibility, high manufacturing efficiency, exceptional safety profiles, and broad potential in the medical field. These novel vaccines necessitate stringent quality control measures to guarantee product stability and safety.
Mature mRNA primarily comprises five structural components, initiating from the 5' end are the 5'-end cap region, the 5' untranslated region (UTR), the antigen-encoding open reading frame (ORF), the 3' UTR, and the polyadenine tail (poly(A) tail). The 5'-end cap enhances mRNA stability during translation, splicing, polyadenylation, and nuclear export, renders the mRNA resistant to cytolytic enzymes in vitro and extends its half-life. The structural and functional properties of the 5'-end cap influence protein synthesis and, consequently, the expression of antigens that can elicit an immune response. The rate of 5' capping of mRNA serves as an indicator of mRNA stability.
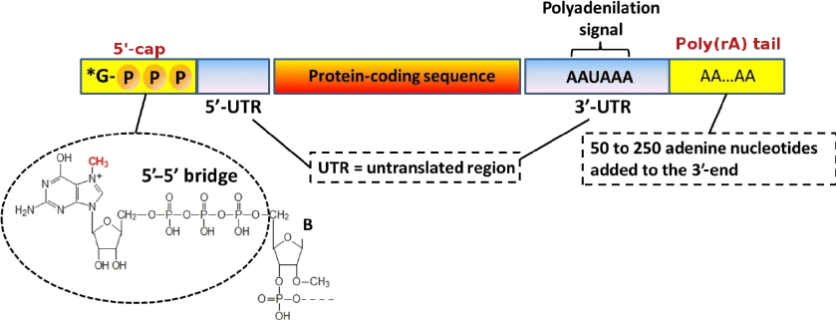
Figure 1. Capping of the 5' End of mRNA
In recent years, mass spectrometry has been leveraged for the analysis of mRNA 5' capping efficiency, notable for its high throughput and sensitivity. MtoZ Biolabs offers an extensive service for mRNA 5'-capping rate analysis, enabling detailed assessments for mRNA vaccines, in vitro mRNA, among others.
A key process in the processing of mRNA precursors is the addition of a 7-methylguanosine (M7G), also known as a 'Cap', to the 5' end of the mRNA. Capping enhances mRNA stability and provides protection against degradation by ribonucleases.
The capping step yields a range of structures, with the most prevalent being the “CAP0” structure, characterized by methylation at the N-7 position of the guanine ring. The structure is termed “CAP1” when methylation extends to the ribose of the adjacent nucleotide, and “CAP2” when the methylation occurs on the 2'-hydroxyl group of the ribose. Furthermore, during the dephosphorylation process, a variety of related impurities emerge, including monophosphoric acid, diphosphoric acid, and triphosphate.
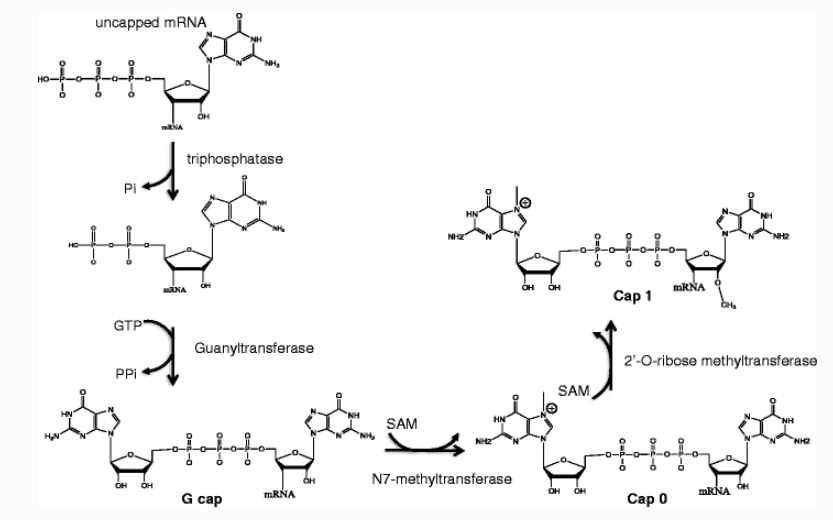
Figure 2. Different 5' Capping Structures and Impurities in the Dephosphoric Acid Process
The basic principle of mRNA capping rate analysis is that the short capping chain at the 5' end is isolated following enzymatic pretreatment and separation of the sample, and mass spectrometry allows for the precise characterization of the molecular weight of the short capping chain, enabling the differentiation of structures with varying molecular weights that form during the capping process, and the capping rate is determined through the quantitative analysis of various structures.
Analysis Workflow
1. Sample Pre-Processing
Using RNAse digestion combined with magnetic bead separation method to obtain 5'-end capped short chain.
2. Mass Spectrometry Data Acquisition
Liquid Chromatography separation of different 5'-end capped structure, to collect different 5'-end capped structures' mass-charge ratio information.
3. Data Analysis
The relative abundance and ratio of different 5'-capped structures were determined by qualitative analysis and peak area quantitative analysis of different 5'-capped structures using special mass spectrometry software.
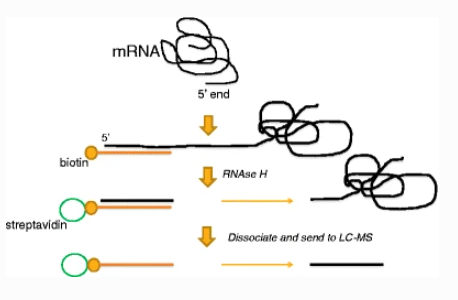
Figure 3. mRNA 5' Capping Rate Analysis Process
Services Advantages
1. High-precision mass spectrometer ensures precision and accuracy. The Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer can separate isotopic peaks from the baseline, thereby obtaining precise molecular weight information of single isotope.
2. Results reports are provided within two weeks.
3. It has a wide range of applications and supports the common quantitative analysis of various mRNA 5'-capped structures.
Sample Results
1. Experimental Procedures
2. Related Mass Spectrum Parameters
3. Raw Data
4. The Total Ion Chromatogram (TIC) of mRNA 5'-Capped Structure
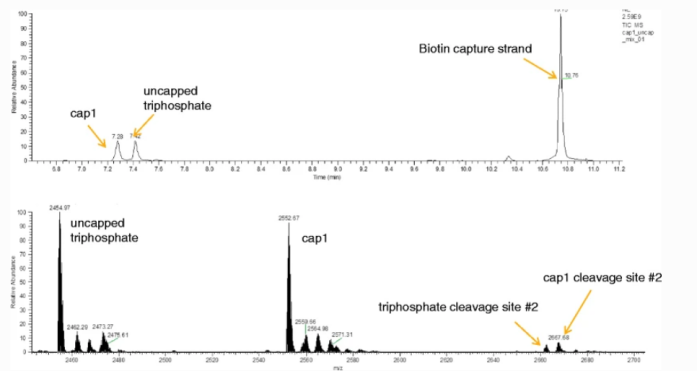
Figure 4. TIC Diagrams of Different 5'-Capped Structures
5. Type of mRNA 5'-Capped Structure and 5'-Capped Rate Table
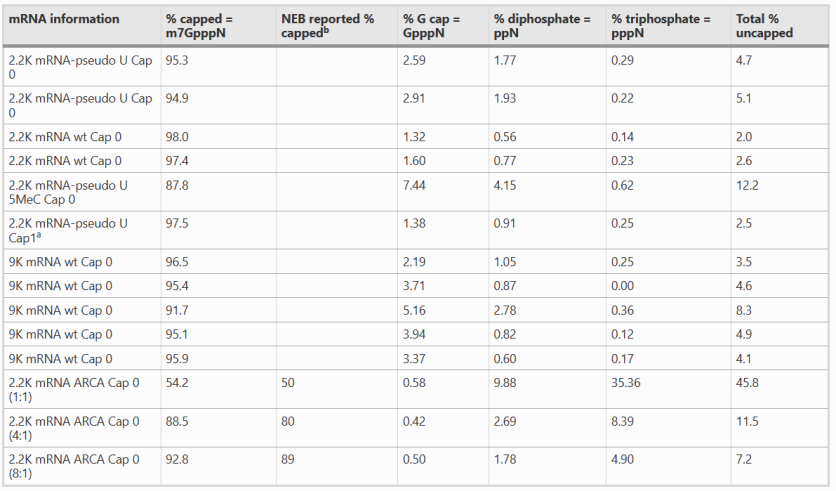
Figure 5. Capping Type and Capping Rate
Sample Submission Requirements
1. Purified capped mRNA samples are accepted.
2. The types of mRNA 5' capped structures that could be detected include Cap0, Cap1, and Cap2.
Services at MtoZ Biolabs
1. mRNA 5' Cap Structure Type Determination and Quantification
2. mRNA 5' Capping Rate Assay
Applications
1. Quality Control of mRNA Vaccine
2. mRNA Stability Analysis
FAQ
Q1: What are the functions of the mRNA 5' end cap?
The 5' cap enhances mRNA stability during translation, splicing, polyadenylation, and nuclear export, imparts resistance to cytolytic enzymes in vitro-transcribed mRNA, and extends mRNA's half-life. The structural and functional attributes of the 5'-end cap influence protein biosynthesis, thereby modulating antigen expression capable of triggering an immune response.
Q2: What are the advantages of high-resolution mass spectrometry analysis of mRNA sequence structure compared to next-generation sequencing?
High-resolution mass spectrometry enables rapid and accurate analysis of RNA sequences, as well as in-depth characterization of the types, sites, and content of post-translational modifications. Furthermore, it can also be used for both qualitative and quantitative analysis of RNA metabolites.
How to order?







