Metaproteomics Service
- Microbiome analysis in human health and disease.
- Environmental microbiome studies (soil, water, air).
- Bioremediation and microbial community monitoring.
- Industrial applications like fermentation and bioprocessing.
- Agriculture and soil health assessments.
- Biodiversity and ecosystem function analysis.
Metaproteomics is an powerful approach that integrates proteomics with microbial community analysis, enabling the identification and characterization of proteins directly from complex microbial ecosystems. Unlike traditional proteomics, which typically focuses on individual organisms or isolated cells, metaproteomics examines the collective protein expression of entire microbial communities, including bacteria, fungi, archaea, and viruses. This technique provides a detailed snapshot of microbial function, metabolic activities, and interactions within their native environments. Metaproteomics approach is particularly valuable in understanding the functional roles of microorganisms in environments such as the human gut, soil, or aquatic systems, where the interactions between microbes play critical roles in health, nutrient cycling, and ecosystem stability.
The applications of metaproteomics span a wide array of scientific disciplines, with profound implications for both basic and applied research. In environmental microbiology, metaproteomics is used to explore the complex interactions between microbial populations and their environments, whether in soil, water, or extreme habitats. By identifying which proteins are being expressed, scientists can gain insight into how microbes respond to environmental stressors, nutrient availability, or changes in the ecosystem. In human health, metaproteomics is increasingly employed to study the microbiome, particularly the gut microbiota, which plays a pivotal role in digestion, immunity, and disease. By analyzing the protein profiles of microbiota, researchers can uncover biomarkers associated with diseases such as inflammatory bowel disease (IBD), obesity, and even mental health disorders. Metaproteomics also offers significant potential in industrial and biotechnological applications, where understanding microbial functions at the protein level can enhance processes like fermentation, bioremediation, or bioengineering.
Services at MtoZ Biolabs
Despite its great potential, metaproteomics faces several challenges due to the complexity and diversity of microbial communities. A single sample may contain thousands of different organisms, each with distinct protein expressions, making it difficult to identify and quantify proteins accurately. The presence of non-model organisms, lacking complete genomic data, further complicates protein identification. While high-throughput mass spectrometry enables deep proteomic analysis, managing and interpreting the large datasets generated requires advanced bioinformatics tools. Effective metaproteomics demands both cutting-edge technology and expert-driven experimental workflows to ensure precise identification and quantification.
At MtoZ Biolabs, we address metaproteomics challenges by leveraging advanced sample preparation techniques, including protein enrichment and fractionation, to reduce impurities and enhance protein detection from complex microbial communities. To overcome database limitations, we integrate de novo sequencing and combine metagenomic and metatranscriptomic data to identify novel or poorly annotated proteins. This multi-omics approach enables a comprehensive understanding of protein function. Additionally, we use functional prediction tools based on sequence homology and structural motifs, ensuring accurate annotation of unknown proteins. Our robust, integrated workflows deliver reliable insights from complex microbial ecosystems.
Analysis Workflow
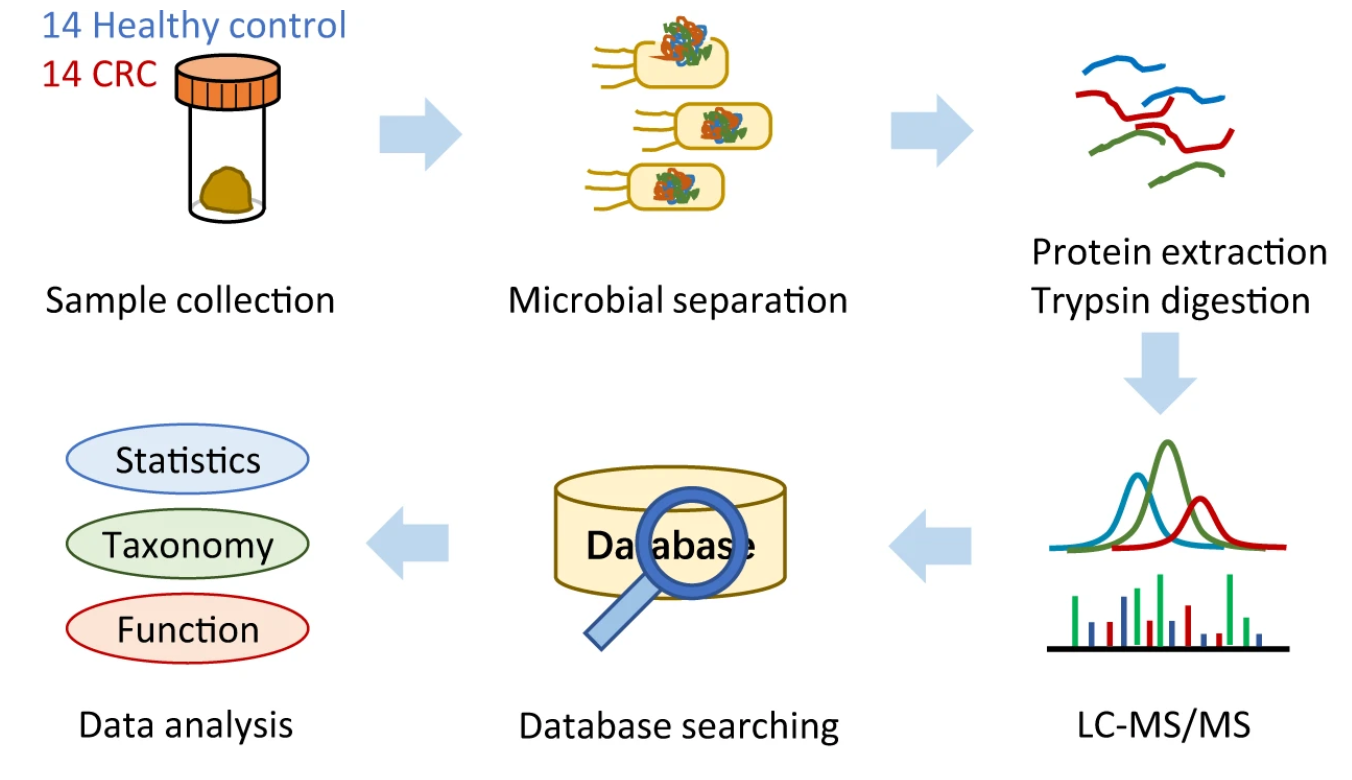
Figure 1. Workflow for Metaproteomics of the Gut Microbiome of Colorectal Cancer (CRC) Patients and Healthy Crowds
Why Choose MtoZ Biolabs?
1. Advanced Analysis Platform
MtoZ Biolabs established an advanced Metaproteomics Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. Optimized Protein Extraction
We offer specialized protein extraction methods optimized for various environmental samples, ensuring high-quality and reproducible results.
3. Advanced Grading Technology
Our use of grading technology provides in-depth analysis, capturing comprehensive metaproteome expression profiles for accurate insights.
4. High-Data-Quality
Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all metaproteomics data, providing clients with a comprehensive data report.
5. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
Applications
Our Metaproteomics Service provides in-depth analysis of microbial communities for diverse applications.
Case Study
1. A Metaproteomics Study of the Bering Strait Microbiome
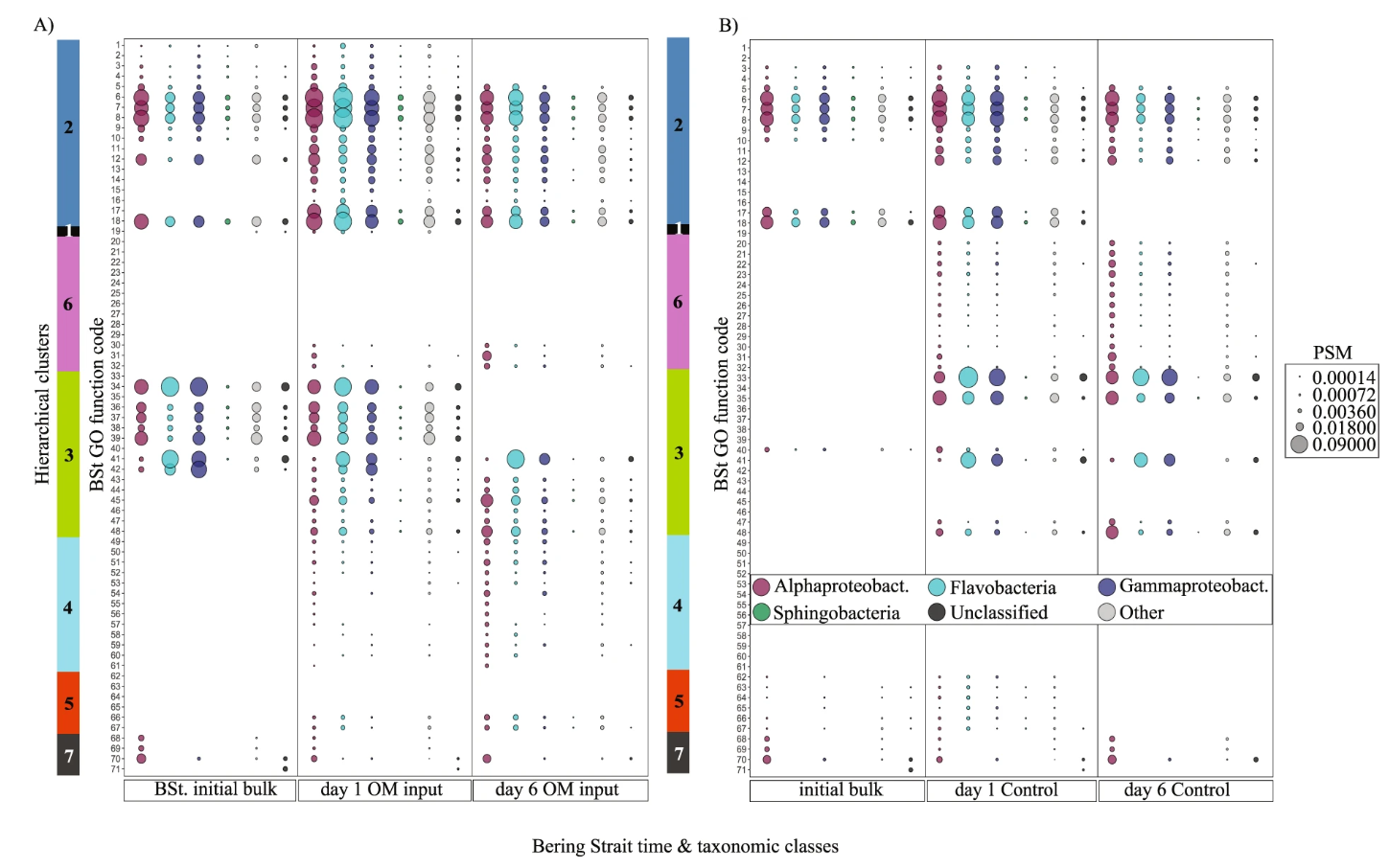
Mikan, M. P. et al. ISME J. 2020.
Figure 2. Peptide Spectrum Match (PSM) Values for the Six Major Bering Strait (BSt) Microbiome Taxonomic Categories and GO Functional Changes
2. A Metaproteomics Study of Human Gut Microbiome
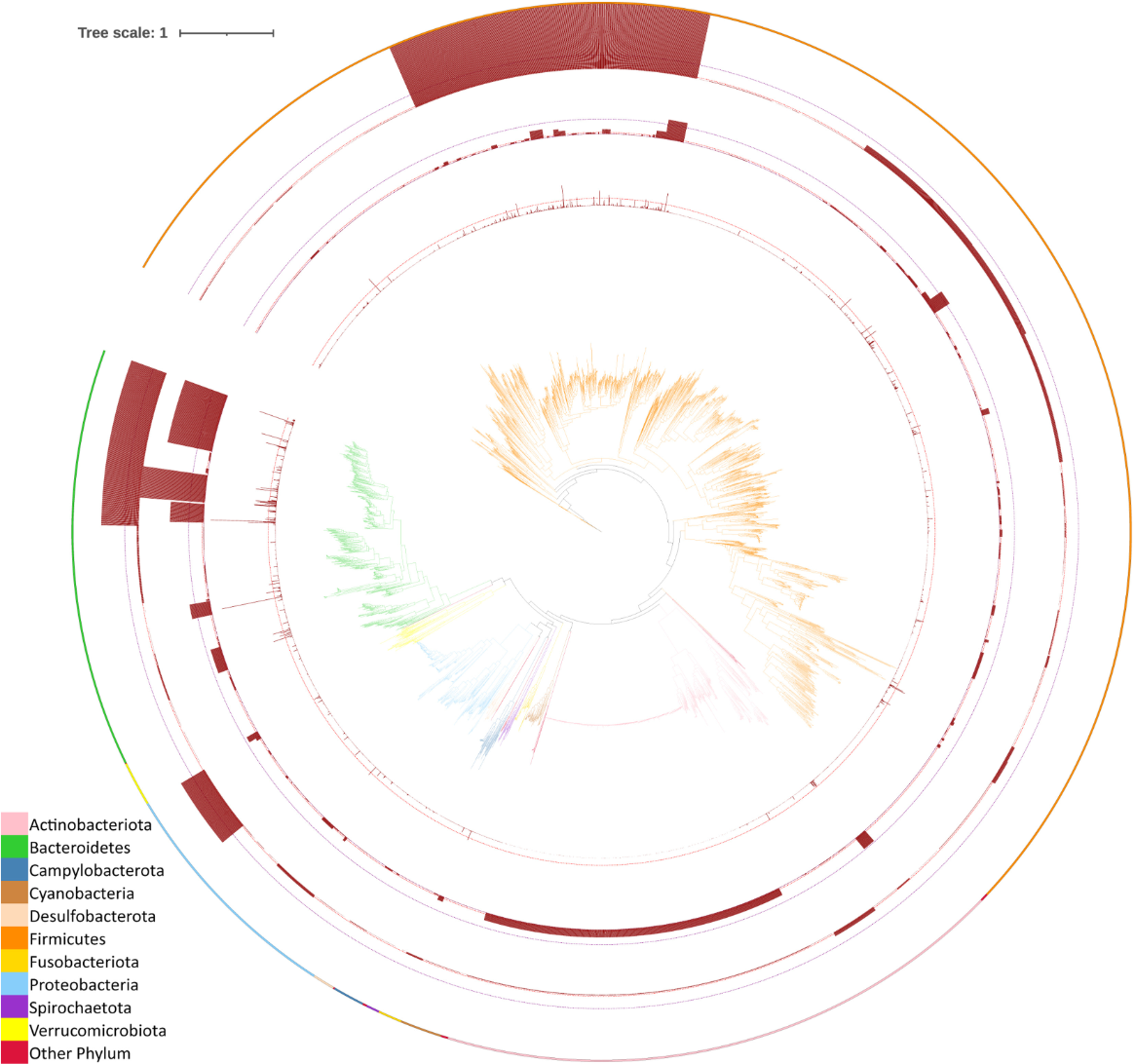
Sun, Z. et al. Mol Cell Proteomics. 2024.
Figure 3. Phylogenetic Distribution of Identified Peptides in Human Gut Metaproteomics Studies
FAQ
Q: Is it possible to conduct metaproteomics on species without metagenomic or metatranscriptomic sequencing?
For species that lack sequencing data, you can use gene sequences from closely related species as a reference database for proteomics studies. When closely related species are not available, broader taxonomic groups (such as genera or families) can be used to construct a reference database for protein identification.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our Metaproteomics Service is designed to provide high-throughput, cost-effective solutions with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







