Lung Disease Diagnosis-Applied Exosomes
Lung Disease Diagnosis-Applied Exosomes is a service that utilizes high-purity exosome isolation and analysis workflows to deeply explore the molecular contents of exosomes. By integrating transcriptomic, proteomic, and metabolomic strategies, this service aims to uncover molecular mechanisms related to lung diseases and support the development of preclinical biomarkers.
Exosomes are nanoscale membrane-bound vesicles secreted by various cell types and can be found in multiple biological fluids, including blood, sputum, bronchoalveolar lavage fluid (BALF), and bronchial secretions. The molecular cargo carried by exosomes reflects the physiological or pathological state of their parent cells. During disease onset and progression, exosomes derived from diseased tissues exhibit distinct changes in expression profiles, cargo composition, and secretion levels.
Xu K. et al. Biomedicine & Pharmacotherapy. 2021.
Lung diseases—such as lung cancer, chronic obstructive pulmonary disease (COPD), pulmonary fibrosis, and asthma—are often diagnosed at a late stage due to subtle early symptoms or the invasiveness of traditional diagnostic methods. In recent years, exosomes have gained significant attention as non-invasive biomarkers for lung diseases. As key mediators of intercellular communication, exosomes carry molecules such as proteins, miRNAs, mRNAs, and lipids, which are known for their high stability, broad origin, and ease of detection.
Leveraging an advanced exosome isolation and purification platform combined with multi-omics mass spectrometry technologies, MtoZ Biolabs provides the Lung Disease Diagnosis-Applied Exosomes service by analyzing exosomes and their contents to screen biomarkers that can be used for lung disease diagnosis, helping researchers obtain high-quality, verifiable data support in early disease screening, progress monitoring, and treatment target exploration.
Analysis Workflow
1. Exosome Isolation and Quality Control
High-purity exosomes are isolated using a variety of established methods, followed by comprehensive quality control assessments.
2. Extraction of Target Biomolecules
Target biomolecules are extracted from exosomes according to the specific requirements of each project.
3. Biomarker Detection
Proteomics, metabolomics, or high-throughput sequencing technologies are employed to detect and analyze the extracted biomolecules.
4. Data Analysis and Report
Data are processed and subjected to bioinformatics analysis, including identification of disease-related biomarkers and functional annotation. A detailed experimental report is provided.
Applications
Examples of specific applications for the Lung Disease Diagnosis-Applied Exosomes service include:
Early Diagnosis and Subtyping of Lung Cancer
Screening for subtype-specific molecules (e.g., for NSCLC or small cell lung cancer) based on miRNA or protein expression profiles in exosomes from plasma or sputum.
Mechanistic Studies of Chronic Lung Diseases
Analyzing exosomal cargo from BALF to uncover mechanisms related to inflammatory pathways, immune regulation, and tissue damage.
Progression Prediction and Target Discovery in Pulmonary Fibrosis
Identifying the roles of miRNAs or long non-coding RNAs in regulating genes associated with fibrosis.
Drug Response Prediction and Efficacy Monitoring
Assessing changes in exosomal expression before and after treatment to evaluate response to immunotherapy, targeted therapy, or bronchodilators.
Biomarker Discovery for Pulmonary Infections
Identifying infection-related molecular patterns and host response factors using exosomes isolated from respiratory fluid samples.
FAQ
Q. How are exosomes isolated from biological samples?
Exosomes are typically isolated using methods such as ultracentrifugation, size-exclusion chromatography (SEC), or immunomagnetic bead separation. These approaches help purify exosomes from other components in biological fluids like blood or sputum.
Q. Are the extracted exosomes thoroughly characterized?
Yes. After isolation, exosomes undergo multi-dimensional characterization, including:
Transmission Electron Microscopy (TEM) to observe the characteristic cup-shaped morphology.
Nanoparticle Tracking Analysis (NTA) to evaluate particle size distribution and concentration.
Western blot analysis to detect exosomal markers such as CD63, CD81, and TSG101.
These steps ensure that the isolated material consists of highly pure and intact exosomes, rather than contaminated particles or cellular debris.
Case Study
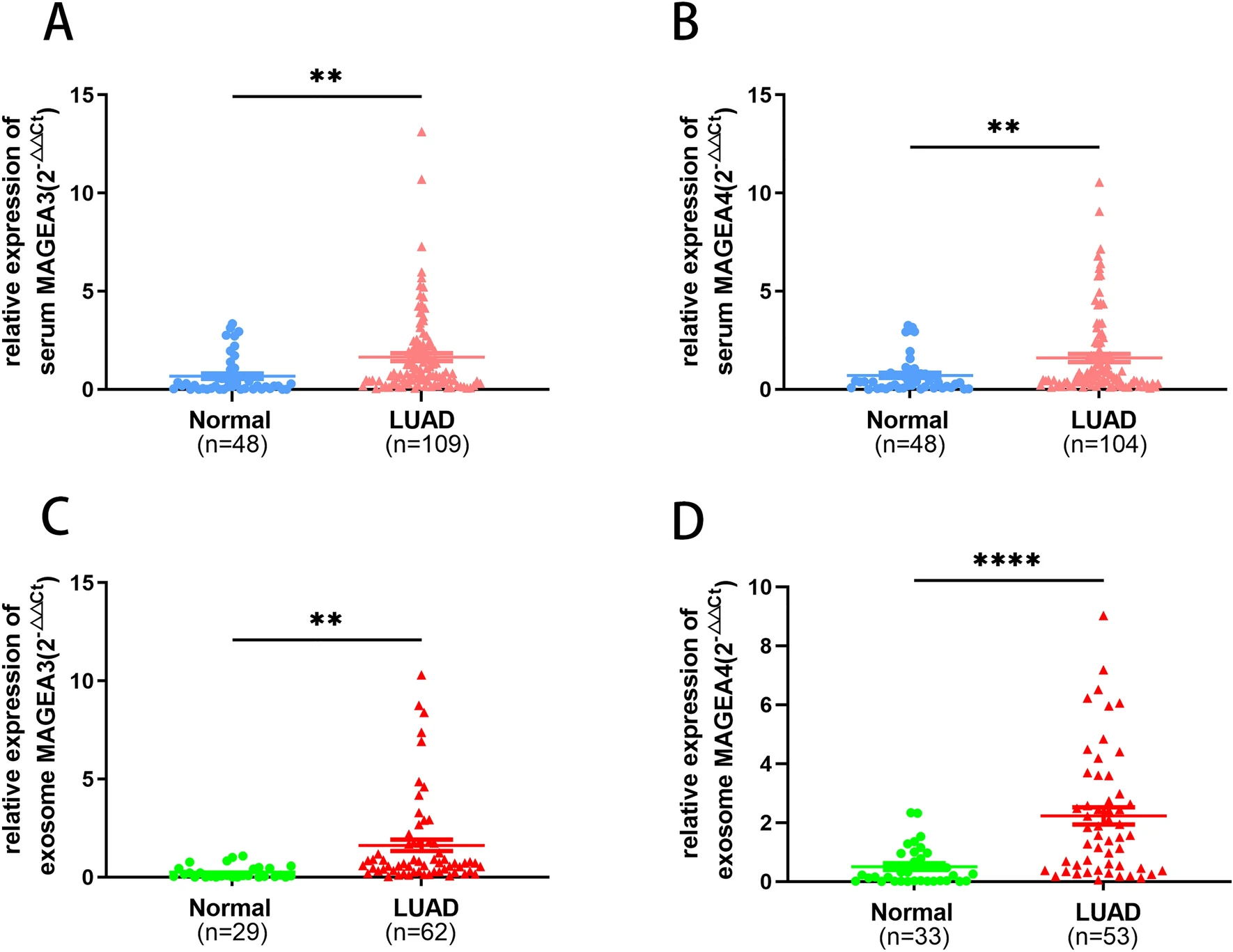
In this study, serum-derived exosomes were isolated using ultracentrifugation and characterized through Transmission Electron Microscopy (TEM), Nanoparticle Tracking Analysis (NTA), and Western blotting for markers CD63 and TSG101. Based on this characterization, researchers used qRT-PCR to assess MAGEA3 mRNA levels in exosomes. Results demonstrated significantly elevated MAGEA3 expression in serum exosomes from lung adenocarcinoma (LUAD) patients, which correlated strongly with tumor size and TNM staging. The biomarker showed an AUC of 0.832, indicating strong potential as a non-invasive early diagnostic marker for LUAD.
Gan Y. et al. Sci Rep. 2024.
How to order?







