Latest Applications of AI in Protein Sequencing
-
DeepNovo: A peptide prediction tool based on Long Short-Term Memory (LSTM) networks, capable of automatically learning ion fragmentation patterns from tandem MS data.
-
PointNovo: Employs a transformer-based architecture to enhance prediction accuracy, maintaining over 80% amino acid-level accuracy even in highly complex sample contexts.
-
Applied to sequence interpretation of proteins from unknown species, novel proteins, and both light and heavy chains of antibodies
-
Addresses the limitations of conventional database matching, particularly in detecting uncharacterized mutations or sequence variants
-
pDeep / PTM-Shepherd: Utilize convolutional neural networks (CNNs) to identify spectral features corresponding to common modifications such as phosphorylation, acetylation, and glycosylation
-
DeepPhospho: Predicts phosphorylation sites directly and provides conservation scores for further evaluation
-
Enables high-resolution identification of modification sites in biotherapeutic antibodies and signal transduction studies
-
Reduces the time required for PTM detection and enhances the degree of process automation
-
AlphaPept: An automated mass spectrometry data analysis platform that integrates peptide identification, quantification, and functional annotation
-
Prosit: Leverages deep neural networks to predict peptide fragmentation spectra, leading to improved identification confidence and consistent quantification
-
Accurately identifies antibody complementarity-determining regions (CDRs) and mutation sites in protein sequences
-
Accelerates large-scale, automated annotation workflows for protein databases
In the current era of AI-driven transformation in the life sciences, artificial intelligence (AI) is redefining the technological framework of protein sequencing. From structural prediction to de novo sequencing, from identification of post-translational modification (PTM) sites to sequence reconstruction, AI technologies are being increasingly integrated into all facets of protein analysis. These advancements not only enhance efficiency and reduce error rates but also empower researchers to better manage the complexity and scale of modern proteomic samples. This paper provides a systematic overview of the latest applications, underlying principles, and leading algorithms of AI in protein sequencing, aiming to support the generation of higher-quality proteomics data.
Three Core Stages of Protein Sequencing Being Transformed by AI
In conventional protein sequencing workflows, three key stages present major technical challenges. The introduction of AI methods has brought significant progress specifically at these critical points:
1. De novo Sequence Prediction
Traditional mass spectrometry-based protein sequencing relies heavily on database searching. However, the advent of AI—particularly deep learning-based neural networks—has made it feasible to achieve high-confidence de novo peptide predictions without the need for reference databases.
(1) Representative Algorithms:
(2) Application Scenarios:
2. Identification and Prediction of Post-translational Modifications (PTMs)
AI models can be trained to recognize the associations between MS/MS fragmentation spectra and specific types of modifications, thereby improving the precision of PTM site identification. These methods are particularly advantageous in samples with coexisting modifications or low signal intensities.
(1) Representative Algorithms:
(2) Application Scenarios:
3. Protein Sequence Reconstruction and Functional Annotation
AI not only facilitates amino acid sequence prediction but also aids in reconstructing full-length protein sequences and performing functional inference based on structural information. This capability is especially valuable in complex tasks such as isoform differentiation and tracking of monomeric or polymeric protein forms.
(1) Representative Algorithms:
(2) Application Scenarios:
Core Advantages of AI in Protein Sequencing
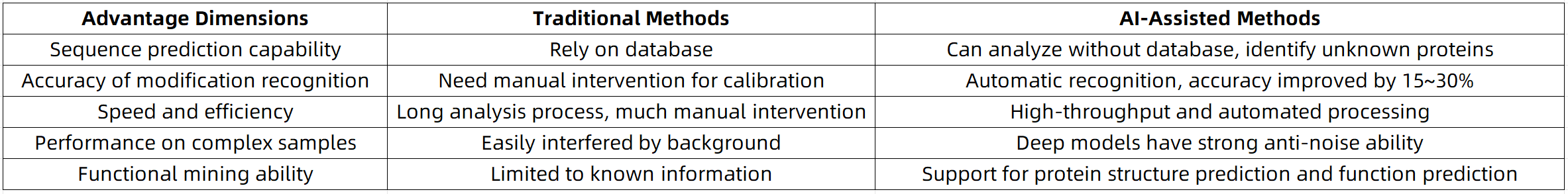
AI not only enhances analytical performance, but more importantly, accelerates the transition of proteomics toward intelligent, high-throughput, and fully automated workflows.
AI + Mass Spectrometry Platform: Intelligent Protein Sequencing Practices at MtoZ Biolabs
As a leading provider of proteomics services, MtoZ Biolabs has integrated AI technologies throughout the entire protein sequencing workflow, establishing an intelligent sequencing platform characterized by high confidence levels and rapid turnaround times. This platform is widely utilized in basic research, antibody drug development, vaccine development, and biomarker discovery. Our AI-driven approaches include:
1. Data analysis through multi-algorithm integration: By combining conventional database search strategies with deep learning models such as Prosit and DeepNovo, we enhance the interpretation of mass spectra.
2. Optimized antibody sequence analysis: We have developed a tailored annotation pipeline that accounts for the structural and functional characteristics of antibody complementarity-determining regions (CDRs).
3. Automated data processing pipeline: A standardized system that converts raw spectral data into finalized analysis reports with minimal manual intervention.
Technological Development Trends
The application of AI in protein sequencing is still in its early stages, yet several significant trends are emerging:
1. End-to-end protein structure prediction: The increasing adoption of tools such as AlphaFold is enabling deeper integration with mass spectrometry data, facilitating automated modeling workflows from linear sequences to three-dimensional protein structures.
2. Cross-omics intelligent integration: AI is expected to serve as a central component in the convergence of proteomic, transcriptomic, and metabolomic data, thereby advancing systems biology research.
3. Single-cell protein sequencing advancements: Improvements in AI algorithms and signal amplification techniques are paving the way for proteome-level analysis at the single-cell scale.
4. Accelerated personalized drug development: AI-driven interpretation of patient-specific protein profiles will support target identification, immunogenicity prediction, and the design of novel therapeutics.
Rather than replacing traditional mass spectrometry methods, AI enhances the interpretability, processing efficiency, and analytical depth of proteomic data—while preserving the rigor of experimental science. For researchers, the value of AI lies in its capacity to reconstruct protein-related phenomena with greater speed, precision, and comprehensiveness. At MtoZ Biolabs, we are not only at the forefront of advanced protein sequencing technologies, but also committed to translating AI capabilities into actionable and reliable scientific solutions. Researchers pursuing projects in protein sequence determination, post-translational modification analysis, or antibody sequence validation are encouraged to engage with our AI-powered precision sequencing services.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







