Label Free Analysis Services
Proteins serve as the functional executors of life processes, with their expression levels and dynamic changes within cells directly influencing an organism's physiological state and the onset and progression of diseases. Label-free quantification technology, based on mass spectrometry principles, achieves accurate protein quantification by comparing the ion intensity ratios between samples and standards. Its core steps include sample preparation, enzymatic digestion, peptide separation, mass spectrometry analysis, and data processing. Label free analysis services are characterized by their efficiency and sensitivity in proteomics research, enabling researchers to precisely measure protein abundance changes and uncover their roles in biological processes.
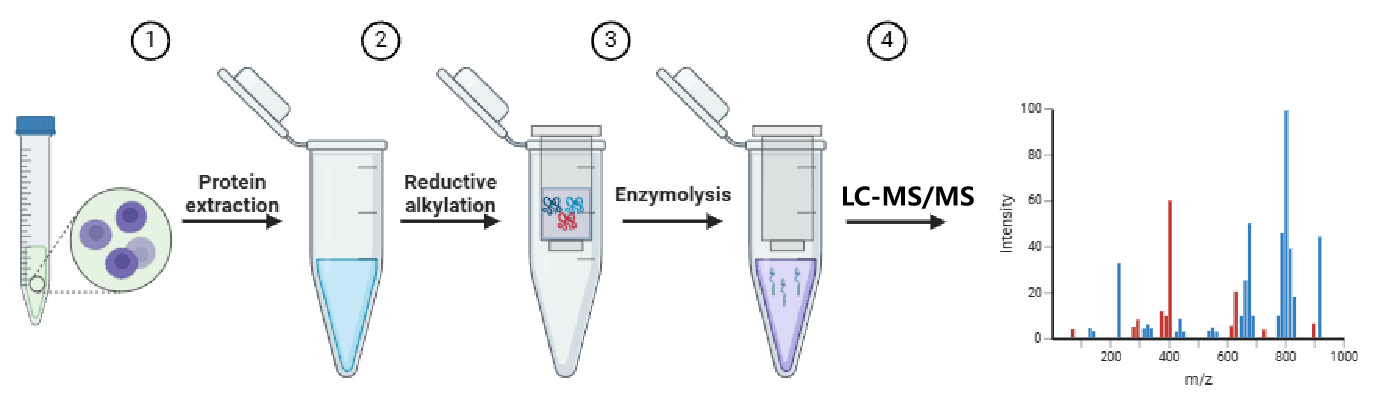
Figure 1. Schematic Diagram of Mass Spectrometry Protein Loading Operation Flow Chart.
Services at MtoZ Biolabs
Leveraging mass spectrometry and imaging technologies, combined with quantitative analysis and high-resolution imaging, MtoZ Biolabs offers label free analysis services to precisely map and analyze the distribution of proteins within cells and tissues. Our services encompass the following two main directions:
1. Label-free quantification (LFQ)
LFQ compares protein abundance based on the ion intensity of peptide segments in mass spectrometry spectra. This technique avoids the limitations of expensive labeling reagents, incomplete labeling leading to sample complexity, challenges in detecting low-abundance peptides, and restrictions on the number of labelable samples. Label-free quantification typically uses precursor ion intensity, where the chromatographic peak area of identified peptides in MS1 represents their relative abundance. The relative expression level of the corresponding protein is then calculated. label free analysis services overcome many limitations of isotope labeling quantification techniques, offering greater flexibility and compatibility with various sample types, enhanced sensitivity for quantifying more proteins, and accurate protein identification down to single amino acid sites. It is a powerful tool for profiling analysis and PTM studies, delivering stable data, excellent reproducibility, broad dynamic quantification range, and high throughput for parallel processing of multiple samples.
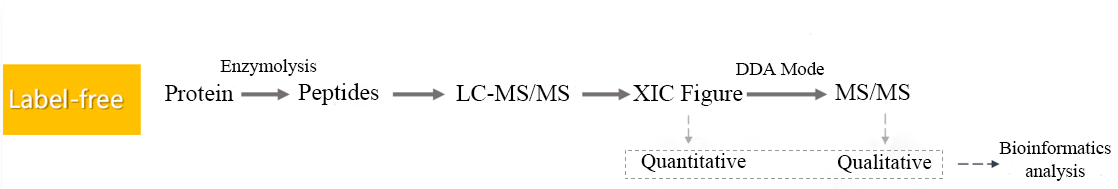
Figure 2. LFQ Quantitative Sample Analysis Process.
2. Data-independent acquisition (DIA)
By continuously scanning all peptide segments, DIA improves quantification accuracy and sensitivity. Traditional acquisition modes in label free analysis services rely on data-dependent acquisition (DDA). In each scanning cycle, DDA selects the top n precursor ion peaks from MS1 for MS2 scanning based on preset criteria, favoring high-intensity peptides while often missing low-abundance peptides. In contrast, DIA is an innovative, comprehensive data-independent acquisition quantification technique. It systematically fragments all precursor ions within specific mass ranges, collecting fragment ion information for all precursor ions to perform protein identification and quantification. DIA does not require predefined target peptides and enables the comprehensive acquisition of fragment information for all ions in the sample without omissions or biases. The scanning points are uniformly distributed, and peptide identification and quantification can be achieved using spectral libraries, ensuring comprehensive and unbiased results.
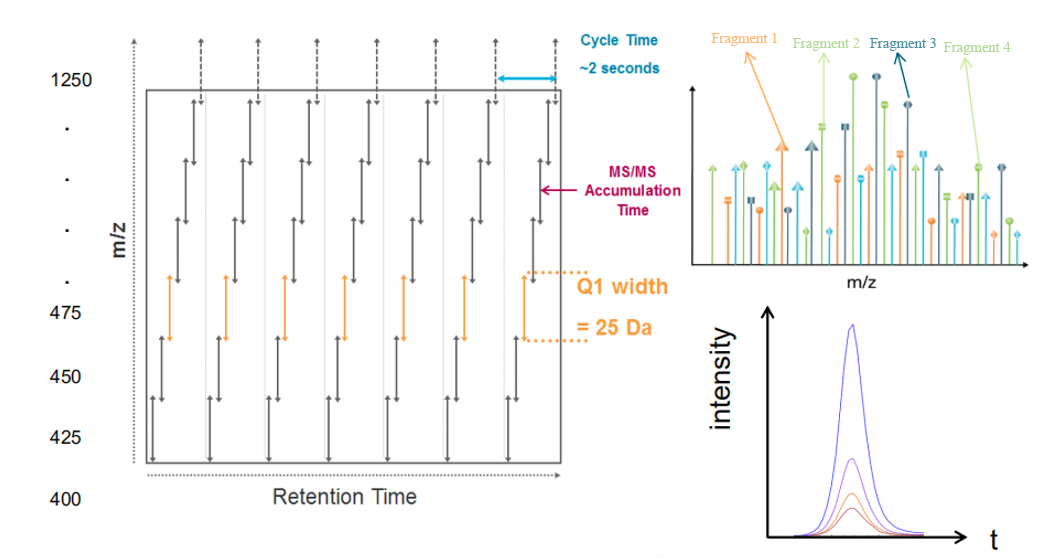
Figure 3. Principle of Quantitative Collection of Label Free DIA.
Service Advantages
1. High Precision and Sensitivity
MtoZ Biolabs leverages Thermo's Orbitrap series mass spectrometers in combination with the Nano-LC system to deliver label free analysis services with exceptional precision and sensitivity, ensuring the accuracy and reliability of quantitative results.
2. Fast and Efficient
Efficient experimental workflows and data processing significantly reduce the experimental cycle time.
3. Professional Team
Our experienced technical team provides expert support and consultation services.
Sample Submission Suggestions
1. Sample Types
We accept a wide range of sample types, including cell extracts, tissue homogenates, serum, plasma, bacterial and fungal cells, and more.
2. Sample Requirements
Samples should be fresh, uncontaminated, and prepared according to specified concentrations and requirements. A minimum of three biological replicates is recommended.
Note: For any special requirements or assistance with sample preparation, please contact us.
Applications
1. Proteomics Research
2. Signal Pathway Analysis
3. Disease Mechanism Research
4. Drug Target Screening
5. Comparative Proteomics
Case Study
1. Engineering Biomimetic Nanoparticles for Targeted Delivery and Efficient Metabolic Synergistic Therapy of Glioblastoma
Glioblastoma multiforme (GBM) is an aggressive brain cancer with limited treatment options. Observing elevated lactate (LA) levels in resected GBM tissues, we developed biomimetic therapeutic nanoparticles (NPs) designed for LA metabolism-based synergistic therapy. Encased in glioblastoma cell membranes, the self-assembled NPs easily cross the blood-brain barrier and target GBM via homotypic recognition.Upon reaching the tumor, the NPs' lactate oxidase converts LA into pyruvate (PA) and hydrogen peroxide (H2O2). PA inhibits cancer cell growth by blocking histone expression and inducing cell cycle arrest. Simultaneously, H2O2 reacts with co-delivered bis[2,4,5-trichloro-6-(pentoxycarbonyl)phenyl] oxalate to release energy utilized by the co-administered photosensitizer chlorin e6, producing cytotoxic singlet oxygen to kill glioblastoma cells.This synergy demonstrated strong therapeutic effects in glioblastoma cell lines and patient xenograft models. This case highlights how label free analysis services confirmed PA's role in blocking histone expression and inducing cell cycle arrest, elucidating disease mechanisms and guiding therapeutic development.
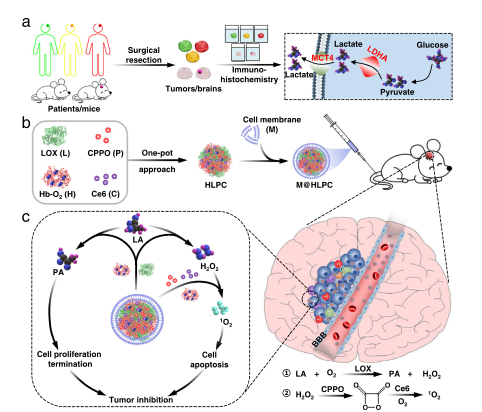
Lu, GH. et al. Nature communication, 2022.
Figure 4. Clinical and Mouse Glioma Analysis and M @ HLPC Tumor Inhibition Construction Schematic Diagram.
2. Integrated Proteomics and Physiological Analysis Reveals the Mechanisms of Interaction Between New Leaf Growth and Storage Protein Degradation in Mature Leaves of Evergreen Trees
This study combines label-free quantitative proteomics with physiological analysis to describe the spatiotemporal mechanisms of new leaf growth and storage protein degradation in mature leaves of citrus trees exposed to low- and high-nitrogen fertilization in the field. Time-series analysis using label-free quantitative proteomics revealed that mature autumn leaves might serve as sites for storage protein synthesis, while aspartic protease proteins are associated with the degradation of storage proteins in mature citrus leaves. Furthermore, bioinformatic analysis of protein-protein interactions indicated that glutamine synthetase and ATP-citrate synthase are key hub proteins in nitrogen remobilization in mature citrus leaves. This case demonstrates how MS-based label free analysis services provide detailed insights into protein dynamics, elucidating the mechanisms underlying the interaction between new leaf growth and storage protein degradation in mature leaves of evergreen trees.
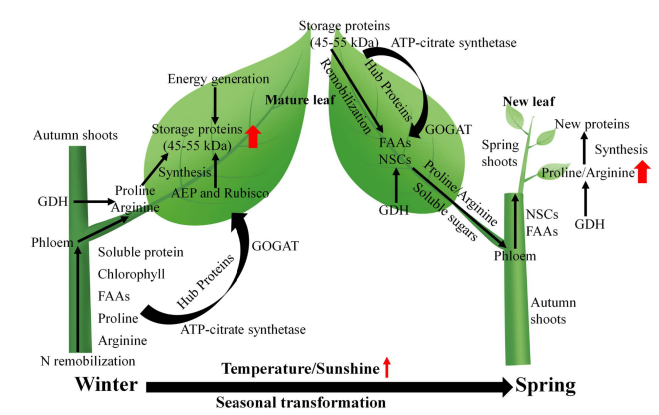
Xiong, H Y. et al. Tree Physiology, 2024.
Figure 5. Pattern Diagram of Winter Citrus Mature Leaf Storage Protein Synthesis, Spring Storage Protein Degradation, and New Shoot Growth.
Deliverables
1. Comprehensive experimental details
2. Materials, instruments, and methods
3. Total ion chromatograms, peptide fragmentation MS/MS spectra, and quality control evaluations
4. Data analysis, preprocessing, and estimation
5. Bioinformatics analysis
6. Raw data files
MtoZ Biolabs offers professional and efficient label free analysis services to support your research endeavors. We look forward to collaborating with you to create a brighter future! Related services include: proteomics analysis, metabolomics analysis, gene expression analysis, bioinformatics analysis, spatial proteomics, multi-omics integration analysis, and customized experimental design. Feel free to contact us for more details!
How to order?







