Intact Protein Analysis Service
Intact Protein Analysis Service utilizes high-resolution mass spectrometry to directly analyze intact protein molecules without enzymatic digestion. Compared to traditional peptide-based analysis, intact protein analysis preserves the native state information of proteins, providing a comprehensive characterization of molecular weight, post-translational modifications (PTMs), isoform distribution, and assembly state. This approach is particularly suitable for the characterization of complex proteins, isomer differentiation, and quality control analysis.
The core principle of Intact Protein Analysis involves directly introducing intact proteins into the mass spectrometer and determining their molecular weight and isotopic distribution using high-resolution mass spectrometry techniques such as Orbitrap and QTOF. By applying deconvolution algorithms, the precise molecular weight of the protein is accurately resolved. Additionally, liquid chromatography-mass spectrometry (LC-MS) enables efficient separation and identification of intact proteins. Furthermore, advanced fragmentation techniques such as higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) can be utilized to analyze PTMs and structural characteristics of proteins.
Leveraging high-resolution mass spectrometry platform, MtoZ Biolabs offers Intact Protein Analysis Service to accurately determine protein molecular weight and assess the relative abundance of protein isoforms. Our service enables researchers to gain in-depth insights into protein characterization, facilitating breakthroughs in protein research and structural analysis.
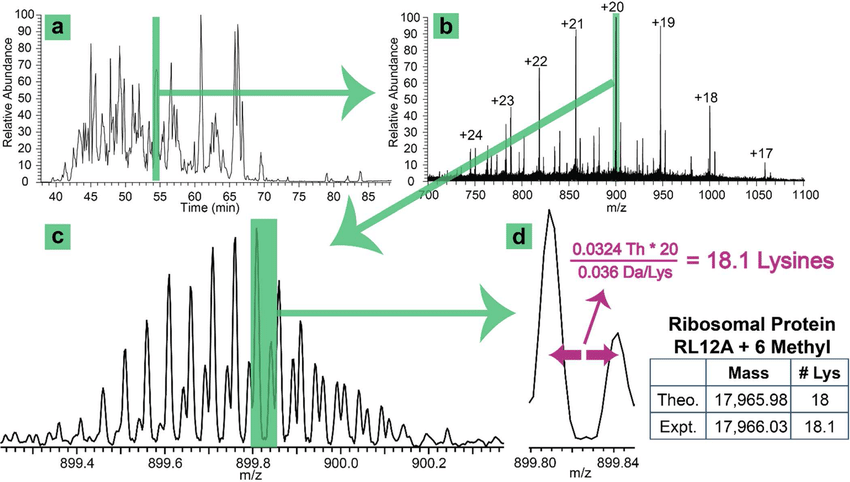
Shortreed M R. et al. Journal of Proteome Research. 2016.
Analysis Workflow
1. Sample Preparation
Optimize sample processing conditions based on sample type and ensure intact protein extraction and preservation.
2. Mass Spectrometry Detection
Analyze intact proteins using liquid chromatography-mass spectrometry (LC-MS) for efficient separation and detection.
3. Data Analysis
Utilize advanced mass spectrometry deconvolution tools to determine precise molecular weight, PTMs, and isoform distribution.
4. Results Analysis & Reporting
Generate a comprehensive report summarizing molecular weight determination, PTMs distribution, and isoform profiling based on high-quality experimental data.
Applications
Examples of Intact Protein Analysis Service applications:
Structural and Functional Studies
Precisely characterize the primary structure, molecular weight, and PTMs of complex proteins.
Drug Development and Quality Control
Intact Protein Analysis Service can be used for molecular weight determination, isoform identification and PTMs analysis of drugs such as monoclonal antibodies, fusion proteins and enzymes to support drug development and manufacturing.
Biomarker Research
Analyze protein integrity, isoform distribution, and modification characteristics in disease-related proteins, providing critical data for biomarker discovery and validation.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Intact Protein Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Intact Protein Analysis data, providing clients with a comprehensive data report.
4. No Need for Protease Digestion:Directly analyze intact proteins without enzymatic digestion, preventing information loss during the digestion process and ensuring comprehensive results.
FAQ
Q. How to Ensure Protein Sample Integrity and Prevent Degradation or Denaturation from Affecting Experimental Results?
Protein integrity is crucial for successful Intact Protein Analysis and the following strategies can help protect protein samples:
1. Sample Storage: Store protein samples at -80°C to prevent degradation and avoid repeated freeze-thaw cycles. Use protease inhibitors to protect samples from enzymatic degradation.
2. Buffer Selection: Use MS-compatible buffers (e.g., low-salt or volatile buffers like ammonium bicarbonate). Avoid buffers with high salt concentrations, detergents, or non-volatile additives that could interfere with MS analysis.
3. Fast Handling: Minimize sample processing time to prevent temperature-induced denaturation. If necessary, perform all handling steps on ice.
4. Quality Check: Before MS analysis, verify protein integrity using SDS-PAGE or HPLC to ensure the absence of degradation bands or impurities.
Q. How to Perform Deconvolution on Complex Mass Spectrometry Data to Accurately Determine Protein Molecular Weight and Modification Status?
1. Software Selection: Use specialized deconvolution software (e.g., BioPharma Finder, MaxEnt, or Protein Metrics) to process complex multi-charge spectra and generate a single molecular weight distribution.
2. Parameter Optimization: Adjust deconvolution parameters (e.g., molecular weight range, isotopic peak width, noise threshold) to ensure precise analysis while removing background noise.
3. Modification Detection: Compare precise molecular weights to identify post-translational modifications (PTMs) such as phosphorylation, glycosylation, or acetylation.
4. Result Validation: Cross-check findings using database searches (e.g., UniProt) and bioinformatics tools. Additionally, confirm modifications with complementary techniques like HPLC-MS or Western Blot.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized Intact Protein Analysis to determine the molecular weight of programmed cell death protein 1 (PD-1). The results revealed a previously unreported O-glycosylation modification in the stalk region of PD-1, leading to a significant change in its molecular weight. Through deconvolution analysis, researchers successfully identified the specific glycosylation sites and confirmed the presence of this modification across different cell sources and physiological conditions. The study suggests that this glycosylation modification may influence PD-1 conformation, stability, and ligand interactions, providing novel insights into immune checkpoint regulation. Furthermore, these findings highlight a potential new target for developing glycosylation-based immunotherapy strategies.
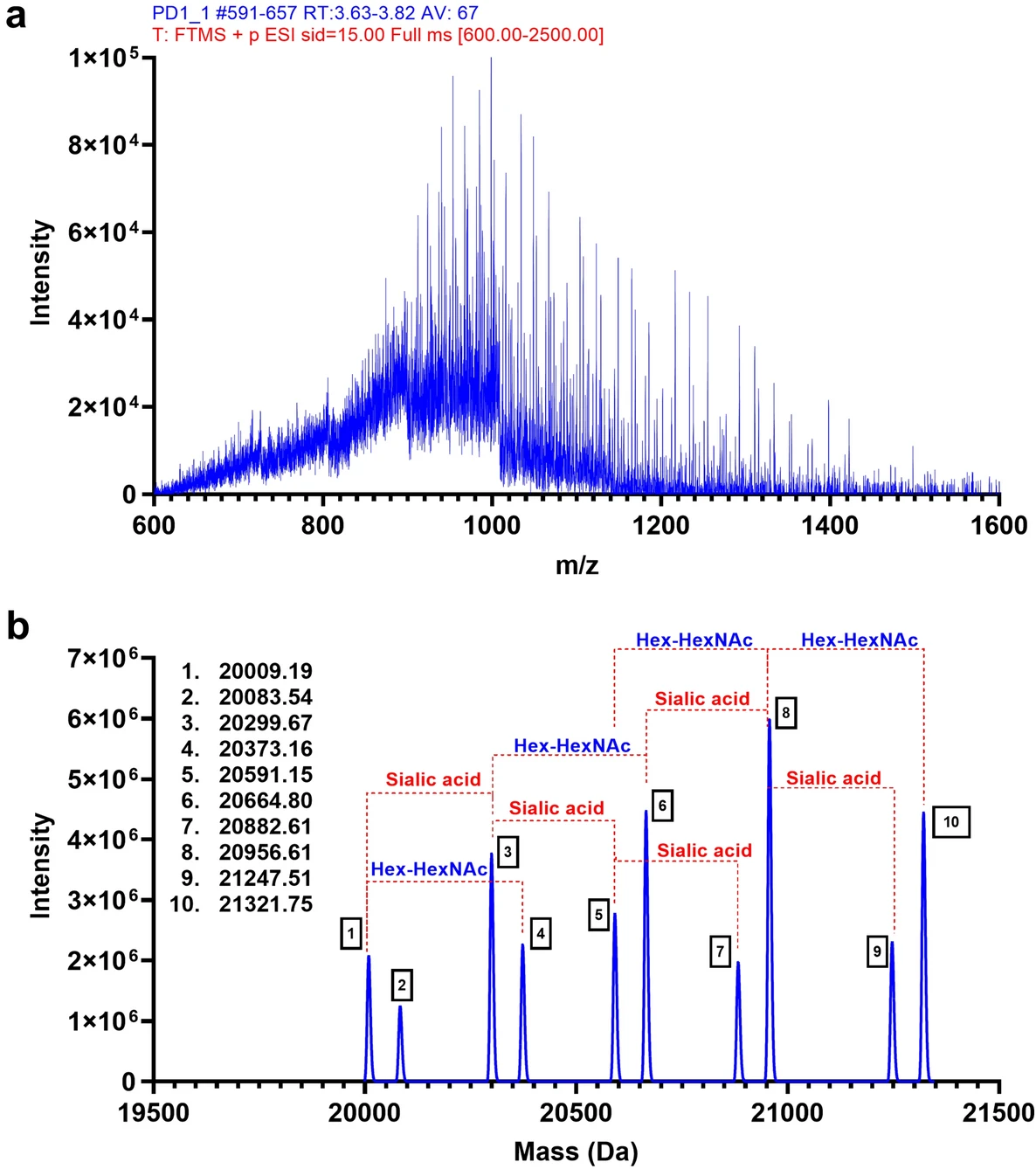
Tit-Oon P. et al. Scientific Reports. 2023.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







