How to Use De Novo Sequencing for Antibody Structure Analysis?
-
Full-length antibody sequence reconstruction (including both variable and constant regions)
-
High-coverage CDR mapping and 3D modeling
-
Annotation of post-translational modification sites and differential analysis
-
Design of recombinant expression constructs
-
Homology-based database comparison and functional annotation
De Novo sequencing refers to determining amino acid sequences directly from protein samples based on MS/MS fragment ion data acquired using high-resolution mass spectrometry, without relying on any reference databases. The core of this technique lies in digesting the protein into numerous short peptides that collectively cover the full sequence, interpreting the corresponding fragment spectra with specialized algorithms, and assembling these sequences to reconstruct the full-length light and heavy chains of antibodies. Unlike database-dependent approaches, De Novo sequencing enables complete sequence determination directly from proteins, making it particularly suitable for complex cases such as antibodies from natural sources, immunized animals, clinical specimens, or historical samples lacking gene vectors.
Why Is Sequence Information Essential for Antibody Structure Analysis?
As highly specific functional biomolecules, antibodies derive their antigen-binding properties, pharmacological mechanisms, safety profiles, and developability from their primary structure—the amino acid sequence. Thorough structural characterization is foundational to antibody drug development, vaccine design, functional reengineering, expression system optimization, and consistency assessment. However, researchers and industry professionals often encounter antibody samples that lack genetic information—such as naturally derived antibodies from immunized animals, legacy samples without expression vectors, or antibodies directly isolated from clinical fluids. In such cases, conventional mRNA-based or database-driven sequencing methods become inapplicable, necessitating mass spectrometry-based De Novo sequencing for comprehensive structural elucidation.
What Is the Workflow of De Novo Antibody Sequencing?
1. Sample Preparation
Purified antibody proteins are required, including SDS-PAGE bands, affinity-purified products, or concentrated supernatants. MtoZ Biolabs offers sample preparation services such as Protein A/G enrichment, desalting, and concentration.
2. Multi-Enzyme Digestion and Peptide Generation
Antibody samples are digested in parallel using multiple enzymes (e.g., Trypsin, Chymotrypsin, AspN) to generate overlapping peptide fragments and maximize sequence coverage.
3. Mass Spectrometry Analysis (LC-MS/MS)
High-resolution mass spectrometers (e.g., Orbitrap Fusion Lumos) are used to acquire MS/MS spectra of peptide fragments, providing detailed ion maps for sequence reconstruction.
4. De Novo Sequencing and Assembly
Dedicated software tools (e.g., PEAKS, Novor) are employed to infer peptide sequences and computationally assemble them into full-length light and heavy chain sequences.
5. Structural Validation and Functional Modeling (Optional)
Follow-up analyses may include structural modeling, expression validation, or epitope mapping to support functional characterization or antibody engineering.
Challenges and Strategies in Antibody Structure Elucidation
Antibodies are characterized by long sequences, structural complexity, and highly variable regions such as complementarity-determining regions (CDRs), posing significant challenges for De Novo sequencing. CDRs often have limited protease cleavage sites and stable conformations, making them difficult to digest completely, which can lead to gaps in sequence coverage. Thus, employing a multi-enzyme digestion strategy is critical to generate diverse, overlapping peptides that improve identification confidence. Additionally, high sequence homology between light and heavy chains can lead to misassemblies during reconstruction, which necessitates the use of structural template alignment and manual curation. Moreover, naturally occurring post-translational modifications (e.g., glycosylation, oxidation, deamidation) can alter mass spectra and cause signal shifts or fragment loss. These must be accounted for in the algorithmic analysis and further validated through expert review. In summary, successful De Novo sequencing requires not only advanced instrumentation but also a comprehensive strategy encompassing proteolysis design, data integration, structural interpretation, and expert validation.
Common Application Scenarios of De Novo Antibody Sequencing
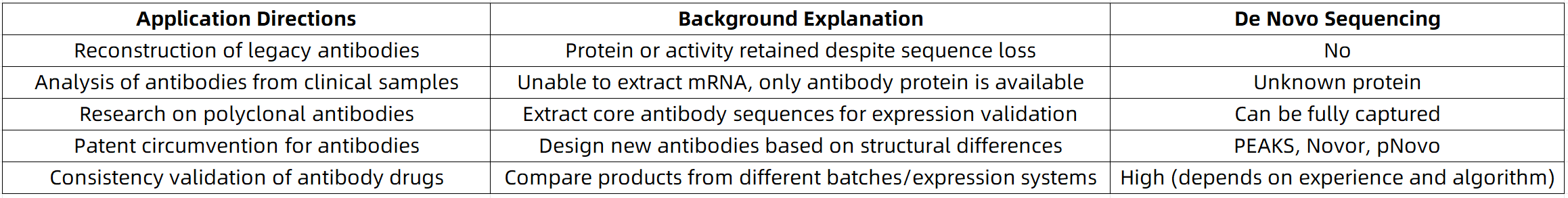
MtoZ Biolabs: Specialized De Novo Antibody Sequencing Solutions
MtoZ Biolabs, a provider of mass spectrometry-based protein structure analysis services, offers a comprehensive workflow encompassing antibody sample preparation, multi-enzyme digestion strategies, high-resolution mass spectrometry, proprietary algorithmic assembly, PTM identification, and recombinant expression validation. Utilizing the Orbitrap Fusion Lumos platform, we conduct parallel enzymatic digestion with five distinct proteases to generate high-coverage peptide matrices. During data analysis, PEAKS software is integrated with our in-house algorithms, further augmented by manual curation and structural modeling to enhance sequence resolution in critical regions such as complementarity-determining regions (CDRs). Our services are tailored for academic institutions, hospitals, and biopharmaceutical companies, offering:
In terms of deliverables, we provide not only the complete antibody sequence but also optional services such as structural modeling, recombinant construct design, and comparative structural analysis with the original antibody. These offerings address diverse requirements ranging from basic research to therapeutic development.
As the pace of antibody innovation accelerates and the demand for data integrity grows, De Novo antibody sequencing is evolving from a specialized technique into a foundational capability. It effectively addresses the structural elucidation of antibodies lacking genetic templates and supports drug consistency verification, functional reconstitution, patent strategy formulation, and expression system optimization. Through a combination of advanced mass spectrometry, rational enzymatic strategy design, and rigorous bioinformatic processing, MtoZ Biolabs is committed to delivering antibody sequence information that is expressible, verifiable, and translatable. Whether you are a research institution or a biopharmaceutical company, we aim to be your trusted partner in antibody structure analysis. If you possess antibody samples without corresponding sequence data, contact MtoZ Biolabs for a free sample evaluation and customized solution. We leverage our technical expertise and extensive experience to support your project from discovery to application.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







