How to Perform High-Precision Antibody Sequencing?
-
Antibody drug development: establishing expression systems, optimizing affinity or stability
-
Antibody humanization: preserving the CDRs while substituting the framework regions
-
Antibody intellectual property applications: ensuring sequence uniqueness
-
Batch consistency analysis: supporting bioequivalence assessments
-
Monoclonal antibody screening and literature-based redevelopment: reproducing published antibodies or validating antigens
-
Standardized multi-enzyme sequencing workflow: a triple digestion strategy employing Trypsin, Glu-C, and Asp-N.
-
AI-assisted sequence reconstruction: dual engines integrating DeepNovo and database search results.
-
CDR identification and framework annotation: automated annotation of antigen-binding regions based on IMGT standards.
-
Comprehensive report output: including amino acid sequence maps, coverage heatmaps, CDR annotations, and confidence scores.
Antibody sequencing has garnered significant attention in recent years as a crucial tool for the development of antibody drugs and the characterization of antibodies. In particular, scenarios such as CDMO contract manufacturing, antibody humanization, patent portfolio design, and consistency analysis have imposed increasingly stringent requirements on the accuracy, completeness, and functional site identification of antibody sequences. How can high-precision antibody sequencing be achieved? What key steps and technical details should be considered during sequencing? This paper provides a comprehensive analysis of the basic principles, mainstream strategies, technical challenges, and solutions of antibody sequencing, offering essential insights into achieving high-quality antibody sequencing.
What Is Antibody Sequencing? Why Is It Important?
Antibody sequencing refers to experimentally determining the amino acid sequence of antibody molecules, with a particular focus on the complementarity-determining regions (CDRs) within their variable regions. The targets for sequencing may include purified antibodies, products expressed by hybridomas, antibodies derived from recombinant expression systems, or natural antibodies produced by B cells.
High-precision antibody sequencing plays a pivotal role in the following tasks:
Mainstream Technical Approaches for Antibody Sequencing
Antibody sequencing methods can be broadly classified into two categories:
1. Antibody Gene Sequencing
This approach involves RNA extraction → reverse transcription → PCR amplification → next-generation sequencing (NGS) → translation into amino acid sequences
(1) Advantages:
High sensitivity with the capability to trace back to individual B cell clones
Applicable to B cell libraries and high-throughput screening scenarios
(2) Limitations:
Provides only predicted theoretical sequences, lacking the ability to detect post-translational modifications or mutations
Inapplicable to antibodies without available cellular sources (e.g., purified antibody products)
2. Mass Spectrometry-Based De Novo Sequencing
This approach involves enzymatic digestion of antibodies → mass spectrometry analysis → peptide identification → de novo reconstruction of full-length protein sequences
(1) Advantages:
Directly analyzes the protein itself, independent of cell or nucleic acid information
Capable of identifying actual expressed products, including modifications, mutations, and isoforms
Suitable for commercial antibodies, antibodies from literature, crude antibody preparations, and diverse sample types
(2) Limitations:
Requires advanced experimental platforms and algorithms; certain structures (e.g., N-terminal blockage) are difficult to resolve
In light of its broad applicability and critical importance for sequence accuracy, high-precision mass spectrometry-based de novo sequencing has become the preferred method in the industry.
Key Technical Aspects of High-Precision Antibody Sequencing
To achieve an accuracy exceeding 98% and a sequence coverage exceeding 95%, it is essential to optimize the following technical components:
1. Multi-Enzyme Digestion Strategy
(1) Employ a combination of specific enzymes (e.g., Trypsin, Glu-C, Chymotrypsin, Asp-N) for enzymatic digestion.
(2) Generate a diverse range of overlapping peptides to avoid regions lacking peptide coverage.
(3) Facilitate crossing domain boundaries, enabling the reconstruction of long sequence fragments.
2. High-Resolution Tandem Mass Spectrometry Analysis
(1) Utilize high-resolution instruments (e.g., Orbitrap Eclipse, timsTOF Pro 2) capable of achieving a resolution exceeding 120,000.
(2) Apply advanced fragmentation techniques (HCD/ETD) to acquire detailed peptide fragment spectra.
(3) Enable detection of peptides with multiple charge states and those present at low abundance.
3. AI-Assisted de novo Sequence Reconstruction Algorithms
(1) Utilize deep learning models (e.g., DeepNovo, Novor, pNovo+) to predict amino acid sequences.
(2) Integrate with database search results to enhance matching accuracy.
(3) Effectively identify non-canonical fragments, post-translational modifications, or mutation sites.
4. Automated Recognition of Antibody Framework and CDR Regions
(1) Leverage databases such as IMGT, Abysis, and IgBlast to align the framework regions and precisely locate CDR1, CDR2, and CDR3.
(2) Distinguish between light and heavy chains and confirm the integrity of the variable regions.
(3) Facilitate analysis of mixed sequences from polyclonal samples.
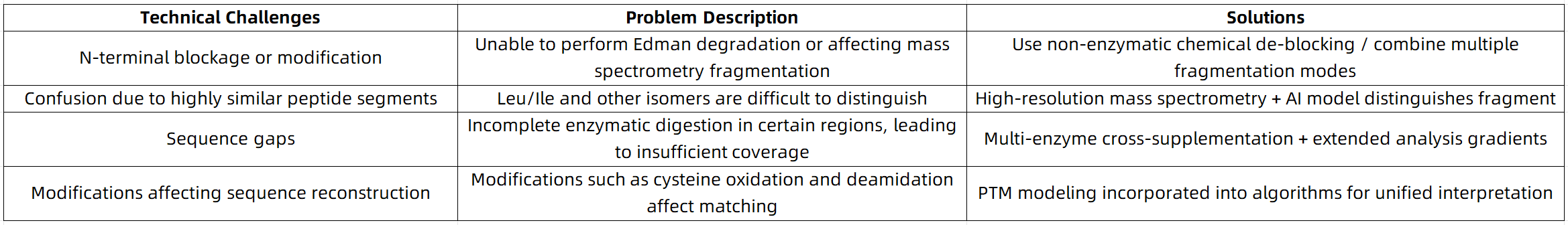
Common Challenges and Solutions in Antibody Sequencing
Advantages of the MtoZ Biolabs Antibody Sequencing Platform
As a professional provider of proteomics and antibody sequencing services, MtoZ Biolabs has established a high-precision sequencing platform optimized for antibody samples, offering comprehensive services covering various sample types such as crude samples, hybridomas, and expressed products:
Sequencing accuracy exceeding 98% and sequence integrity exceeding 95%, supporting hundreds of pharmaceutical companies, CROs, and research institutions in antibody analysis, regulatory submissions, and other projects.
With the continued expansion of the antibody drug market, antibody sequencing is no longer an option but a fundamental requirement for molecular traceability, design optimization, and quality assurance. High-precision antibody sequencing requires not only state-of-the-art mass spectrometry platforms but also relies on the synergy of deep algorithm integration, protein annotation capabilities, and standardized experimental workflows. At MtoZ Biolabs, we focus not only on generating sequences but also on ensuring accuracy, interpretability, and rapid turnaround. For inquiries regarding antibody sequence analysis, translational research, or patent verification, please contact us for professional advice and customized solutions.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







