How to Choose the Right Protein Sequencing Technology?
-
High-resolution mass spectrometry platforms: Orbitrap Eclipse, timsTOF Pro 2
-
Multiplexed enzymatic digestion strategies: Trypsin, Glu-C, Chymotrypsin combinations
-
High-confidence sequence reconstruction algorithms: Supporting both de novo and hybrid database-based approaches
-
Specialized antibody sequencing services: Full workflow characterization of hybridoma cells, antibody liquids, and expression products
-
Expert data interpretation reports: Including sequence maps, confidence scores, alignment results, and intuitive visualizations
Protein sequencing is a fundamental technique for investigating protein structure and function. It plays a critical role in biomedical research and drug development, including target discovery, mechanism elucidation, biomarker identification, and antibody characterization. With continuous advancements in mass spectrometry and chemical sequencing methods, an increasing number of protein sequencing technologies have become available. These technologies differ significantly in their suitable applications, resolution, cost, and throughput. Faced with such a diverse array of options, how can researchers make informed choices regarding the most appropriate protein sequencing technology? This review provides a systematic overview of current protein sequencing technologies from four perspectives: underlying principles, key advantages, typical applications, and selection guidelines—aiming to support efficient and evidence-based decision-making.
Main Types of Protein Sequencing Technologies
Protein sequencing methods can be broadly categorized into two major types: mass spectrometry-based analytical sequencing approaches (Bottom-up and Top-down), chemical sequencing based on Edman degradation, and emerging single-molecule techniques. The commonly employed methods include:
1. Bottom-Up Protein Mass Spectrometry Sequencing (Peptide-Level)
(1) Principle: Proteins are enzymatically digested into peptides, followed by LC-MS/MS analysis to determine peptide sequences.
(2) Advantages: High throughput and sensitivity; well-suited for complex biological samples.
(3) Limitations: May result in incomplete sequence coverage; information on post-translational modifications (PTMs) can be lost.
2. Top-Down Protein Sequencing (Full-Length Level)
(1) Principle: Intact proteins are directly fragmented and analyzed using high-resolution mass spectrometry to reveal their sequence and structural features.
(2) Advantages: Enables retention of PTM information; ideal for characterizing individual proteins and isoforms.
(3) Limitations: Requires exceptionally high protein purity and mass spectrometric resolution.
3. Edman Degradation (Classical Chemical Sequencing)
(1) Principle: Sequentially removes and identifies amino acids from the N-terminus of the polypeptide chain.
(2) Advantages: Provides high sequence accuracy; suitable for analyzing short peptides.
(3) Limitations: Typically limited to peptides shorter than 30 amino acids; not applicable to proteins with blocked N-termini.
4. Single-Molecule Protein Sequencing (Under Development)
(1) Principle: Directly reads individual amino acid sequences using nanopore, fluorescence-based, or optical detection methods.
(2) Advantages: Theoretically capable of achieving single-cell resolution.
(3) Limitations: Largely in the research or early commercialization phase; currently associated with high costs and suboptimal accuracy.
Recommended Protein Sequencing Technologies for Different Application Scenarios
Protein sequencing technologies should be selected based on the specific research objectives. Below are strategic recommendations tailored to common scenarios in scientific research and drug development:
1. Antibody Characterization and Sequence Confirmation
(1) Recommended Technologies: Top-down mass spectrometry or Edman degradation in combination with mass spectrometry
(2) Rationale: Accurate identification of antibody variable regions (CDRs) and light/heavy chain pairings is required, with a high demand for sequencing precision
(3) MtoZ Biolabs Solution: Enables full-length antibody sequencing from crude antibody samples, hybridoma supernatants, or purified products, using optimized enzymatic digestion and high-resolution Orbitrap MS; accuracy exceeds 98%
2. Preliminary Identification of Novel or Unknown Proteins
(1) Recommended Technologies: Bottom-up mass spectrometry sequencing (de novo sequencing combined with database search)
(2) Rationale: Suitable for scenarios without prior sequence information, especially when species-specific databases are lacking or when discovering novel proteins
(3) MtoZ Biolabs Support: Employs a comprehensive strategy combining multi-enzyme digestion, multi-level precursor ion selection, and hybrid algorithms using both target and decoy databases to enhance identification of unknown proteins
3. Sequencing of Protein Post-translational Modification (PTM) Sites
(1) Recommended Technologies: Bottom-up mass spectrometry combined with ETD/HCD tandem MS
(2) Rationale: Required for accurate localization of modifications such as phosphorylation, acetylation, and glycosylation
(3) MtoZ Biolabs Advantages: Offers a range of PTM enrichment approaches, coupled with optimized tandem MS fragmentation techniques to improve modification site coverage
4. Consistency and Stability Analysis of Protein Therapeutics
(1) Recommended Technologies: Top-down sequencing along with high-resolution analysis of protein isoforms
(2) Rationale: Applicable for verifying batch consistency, identifying mutations, and analyzing degradation products
(3) MtoZ Biolabs Platform: Utilizes the Orbitrap Eclipse system with resolution exceeding 240,000, enabling detailed analysis at the protein isoform level
Recommendations for Technology Selection: How to Make Informed and Efficient Decisions
When selecting a protein sequencing technology, consideration should be given to the following five key dimensions:
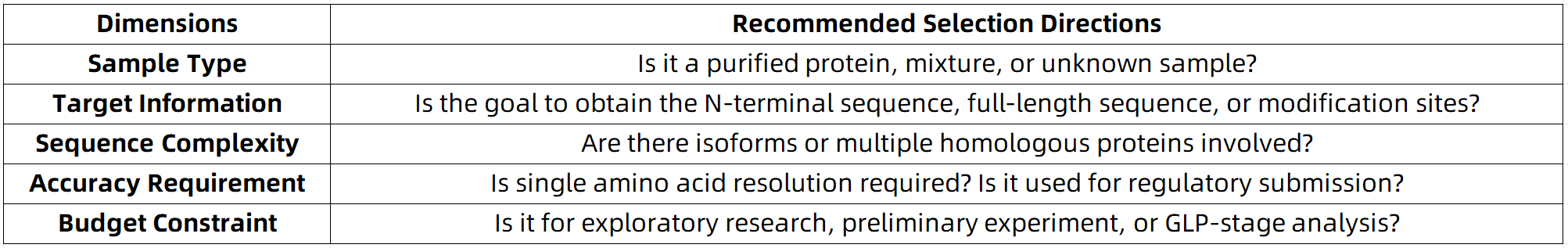
If the appropriate method is not immediately apparent, it is advisable to consult a third-party service provider with integrated multi-platform and multi-technology capabilities—such as MtoZ Biolabs—for comprehensive project evaluation and strategy planning.
Distinctive Features of MtoZ Biolabs’ Protein Sequencing Services
MtoZ Biolabs has established a comprehensive protein sequencing platform, encompassing full-process capabilities from low- to high-throughput analysis, and from standard digestion workflows to advanced post-translational modification detection:
We are dedicated to delivering personalized, one-stop protein sequencing solutions for academic research institutions and biopharmaceutical enterprises. From early-stage discovery to product-level consistency evaluation, our services are designed to facilitate efficient and accurate progression of research and development pipelines.
Protein sequencing technologies are advancing rapidly and continuously extending their applicability in areas such as precision medicine, therapeutic antibody development, and fundamental research. Selecting an appropriate sequencing strategy is critical not only to data quality but also to the efficiency and depth of project execution. If you are currently facing challenges in selecting the most suitable method, we invite you to consult MtoZ Biolabs. We offer complimentary technical evaluations and project planning support to help maximize the impact of your sequencing investment.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







