Comprehensive Host Cell Residual RNA Detection Service
DNA products such as plasmid/minicircle DNA can serve as primary part of DNA vaccine products or as important raw material for viral vectors or DNA vector of gene therapy, and for viral vector or non-viral vector of cellular immunotherapy. Residual RNA in these DNA products may affect biological activity of these products (such as interfering with plasmid transfection and expression efficiency) or pose risks (such as triggering immune responses by activating the cellular interferon pathway due to exogenous RNA), which help establish quantitative detection methods for residual RNA in DNA products.
Regulatory authorities assess the purity of DNA products by considering host cell RNA (HCR) content as a critical quality parameter. In the United States, plasmid quality must include more than 85% supercoiled DNA, less than 1% host cell proteins, less than 1% residual bacterial DNA (PCR detection), and less than 1% residual bacterial RNA (undetectable by high-performance liquid chromatography (HPLC) or gel methods). The European Medicines Agency, in its guidelines for gene therapy medicinal products, recommends evaluating residual HCR to ascertain plasmid purity.
MtoZ Biolabs operates an extensive platform for the analysis of host cell nucleic acids (HCD & HCR) and for the development of nucleic acid detection products. Our services encompass the identification of various host cell DNAs (HCD) and HCR, analysis of HCD fragment lengths and distributions, and the development of corresponding HCD/HCR reagent kits. These capabilities facilitate comprehensive research into nucleic acid extraction, quantitative detection, and fragment analysis. The team can explore optimal experimental conditions through multi-gradient experiments, collect data using various experimental methods such as real-time fluorescent quantitative PCR and capillary electrophoresis, and identify and quantify HCD/HCR through bioinformatics analysis. MtoZ Biolabs offers HCD/HCR extraction, quantification, fragment analysis, and related reagent kit development services, providing a one-stop solution for HCD and HCR analysis.
1. RNA Extraction
Extracting biomolecules like DNA, RNA, and proteins is a fundamental technique in molecular biology. It marks the starting point for downstream processes and product development, including diagnostic kits. DNA, RNA, and proteins can be isolated from any biological material, such as live or preserved tissues, cells, viral particles, or other samples, for analysis or preparation.
RNA is inherently unstable, with a very short half-life once it is extracted from cells or tissues. Naturally occurring RNAs include ribosomal RNA (rRNA) (80%-90%), messenger RNA (mRNA) (2.5%-5%), and transfer RNA (tRNA). RNA isolation must be taken care due to the easy degradation of RNA. RNases are enzymes found in blood, tissues, and most bacteria and fungi in the environment, which makes RNA more unstable. Strong denaturants are used throughout the RNA isolation process to inhibit endogenous RNase activity. RNA extraction relies on good laboratory techniques and RNase-free practices.
(1) Acid Guanidinium Thiocyanate-Phenol-Chloroform Extraction
Ulrich et al. (1977) first mentioned the use of guanidinium thiocyanate in RNA extraction. This method is laborious, so Chomczynski and Sacchi (1987) introduced a single-step technique called acid guanidinium thiocyanate-phenol-chloroform extraction, in which homogenate was extracted with phenol/chloroform at a reduced pH. Guanidine thiocyanate is a chaotropic agent used for protein degradation. The principle at the basis of the method is that RNA is separated from DNA after extraction with an acidic solution containing guanidinium thiocyanate, sodium acetate, phenol and chloroform, followed by centrifugation. Under acidic conditions, total RNA remains in the upper aqueous phase, while DNA and proteins remain either in the interphase or the lower organic phase. Then total RNA be recovered by precipitation with isopropanol.
(2) Oligo(dT)-Cellulose Chromatography for Poly(A) RNA Purification
Poly(A) RNA, the template for protein translation, is present at the 3' end of most eukaryotic mRNAs, accounting for 1% to 2% of total RNA. It can be isolated using affinity chromatography on oligo(dT) cellulose. The poly(A) tail forms a stable RNA-DNA hybrid with short oligo(dT) chains and attaches to various support matrices. Due to the formation of only a few dT-A base pairs, high salts must be added to the chromatography buffer to stabilize the nucleic acid duplex. After washing off non-polyadenylated RNA from the matrix, low-salt buffer is used. This buffer helps disrupt the stability of the double-stranded structure and elutes the Poly(A)RNA from the resin.
Common methods for purifying Poly(A) RNA include oligo(dT) column chromatography and batch chromatography. Column chromatography is typically used to purify large amounts (>25 μg) of Poly(A) RNA from mammalian cells. Batch chromatography is the preferred method for handling small amounts (<50 μg) of total mammalian RNA. This method can also be used when dealing with large RNA samples. Batch chromatography is performed using oligo(dT) cellulose at optimal binding and elution temperatures.
(3) Magnetic Beads Method
Magnetic oligo(dT) beads are alternatives to other oligo(dT) matrices for purifying poly(A) RNA from total RNA samples. Poly(A) RNA is extracted by introducing beads coated with oligo(dT). RNA with Poly(A) tails attaches to the oligo(dT). The beads are then pulled to the bottom of the tube, directly removing mRNA from total RNA. The specially treated beads minimize non-specific binding of other nucleic acids and ensure the purity of mRNA.
2. RNA Quality Control
The purity and integrity of RNA are crucial factors for the overall success of RNA-based analyses. Due to the high instability of RNA, RNA fragments are often present in samples, affecting the results of downstream applications.
(1) OD Measurement
Spectrophotometric techniques, such as NanoDrop™, provide information about RNA quantity and purity (i.e., A260/A280 and A260/A230 values) based on the nucleic acid's ability to absorb UV light at a wavelength of 260 nm. Measurements for quantity and quality using a UV/visible spectrophotometer should be performed at multiple wavelengths: 240 nm (possible contaminants absorption), 260 nm (nucleic acids), 280 nm (proteins), and 320 nm (possible contaminants absorption). An OD260/OD280 ratio of 1.7-1.9 indicates high purity of DNA; ratio below 1.7 suggests protein contamination; ratio above 1.9 suggests partial degradation of DNA or presence of residual RNA. One major advantage of this system is the very low sample consumption, only 1-2 µL, which is particularly important when using precious materials like biopsies in human subjects. An A260/A280 ratio greater than 1.8 is generally considered an appropriate indicator of RNA purity. However, the accuracy of this method is questioned because a value of 1.8 only corresponds to 40% RNA. A260 measurement can be influenced by genomic DNA, which may lead to an overestimation of the actual RNA concentration.
(2) Gel Electrophoresis
The second method of inspection is agarose or polyacrylamide gel electrophoresis, where samples are loaded and nucleic acid fragments are separated by their size. The gel is stained with ethidium bromide or SYBR Green, which binds to nucleic acids but not specific to RNA, and the isolated fragments can be visualized by the excitation of the fluorescent dye. RNA concentration can be qualitatively measured by comparing the fluorescence intensity of RNA bands with that of known RNA standards (usually 28S and/or 18S rRNA). When the ratio of the 28S/18S bands is about 2.0 or higher, the RNA is considered to be of high quality. Although these methods are less costly, they require much processing time and a large amount of RNA. It is also difficult to meet the standard of 2.0 rRNA ratio in practice, especially for RNA from clinical samples. On the other hand, a lower rRNA ratio does not necessarily indicate poor quality, especially if no degradation products are detected in electrophoretic traces.
(3) Microfluidics
As mentioned above, traditional methods are often less sensitive and unspecific for single-stranded RNA, and they are susceptible to interference from contaminants and subjective interpretation. For these reasons, many researchers use microfluidics technology (Agilent 2100) to analyze DNA, RNA, proteins, and cells through specific sample chips. The "RNA Integrity Number" (RIN) algorithm is applied to express results.
RIN is a new tool to eliminate individual interpretation in RNA quality control. It takes into account the entire electrophoretic trace, not just the ratio of 28S/18S rRNA with a very small amount of RNA sample (as low as 200 pg). Size standards used during electrophoresis can estimate the size of RNA bands, and the measurement results seem relatively unaffected by contaminants. Moreover, for routine analysis of large amounts of RNA preparations, this is the most convenient and objective method for assessing RNA quality by far . The RIN value ranges from 0 to 10, with 10 representing the highest RNA integrity. By comparing the peak area of the ladder plot, the RNA fragment of known concentration, and the peak area of an unknown sample, the software is able to estimate not only RNA degradation, but also RNA concentration. Another advantage of this software is its ability to categorize small RNAs or microRNAs. But it can not show information about the purity of the sample.
3. RNA Detection
Detection of HCR can be performed using HPLC or agarose gel electrophoresis. Beyond these methods, RT-qPCR and fluorescence quantification of RNA are also utilized to assess residual HCR in biological products.
(1) RT-qPCR
RT-qPCR has been widely used in the industry because it can achieve highly specific amplification and accurate quantification of residual HCR in the sample to be tested by specific primers and probes. RT-qPCR includes four steps: total RNA extraction, RNA reverse transcription to cDNA, real-time PCR assay for amplification and data analysis.
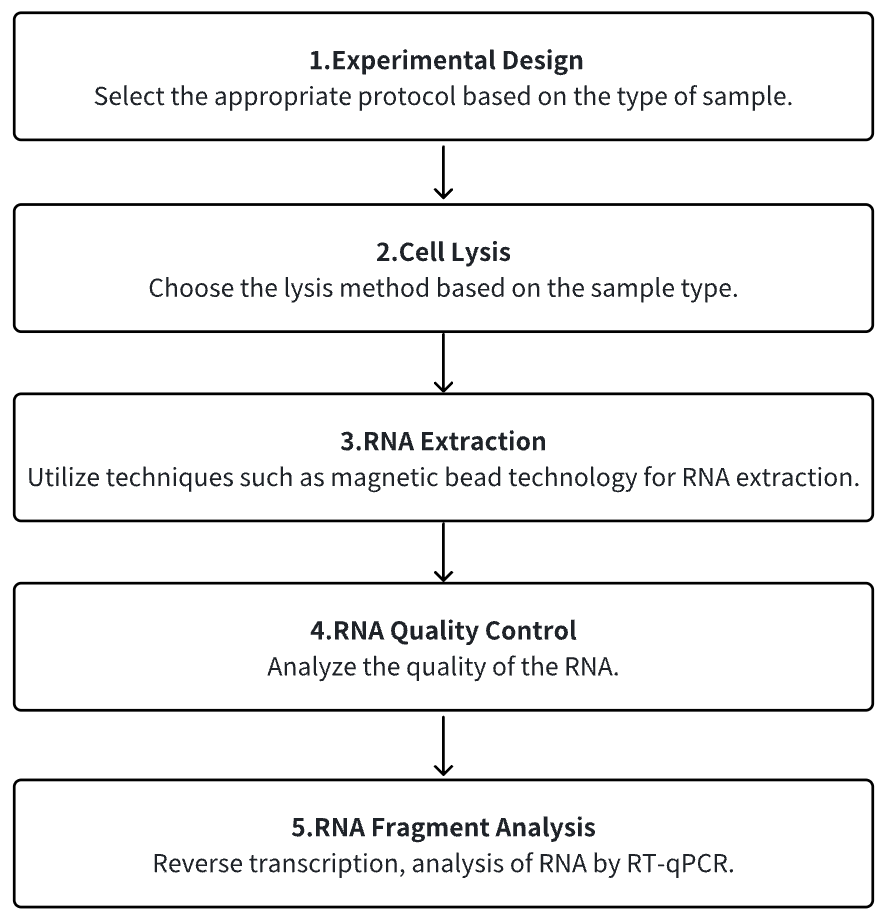
Analysis Workflow
1. Determine the Experimental Protocol Based on Specific Requirements
2. Cell Lysis
3. RNA Extraction
4. RNA Quality Control
5. RNA Reverse Transcription to cDNA
6. RT-qPCR
Service Advantages
1. High Confidence Identification and Characterization of Residual HCR
2. Automate the High-Throughput RNA Extraction Process
3. Effective RNA Detection System and Rich Testing Experience
Example Results
1. Compare RNA Extraction Methods to Face Changes in RNA Quality Using Two Human Biological Samples
Nucleic acids, including RNA, are widely used in biomedicine and biotechnology. Due to their susceptibility to degradation by RNase, the process of handling and extracting RNA from cells and tissues requires professionals and standardized methods to ensure high purity and integrity. Given the variety of technologies available on the market, comparative studies between different RNA extraction methods help researchers or service platforms like biobanks determine the best choice for sample traceability. Studies have compared seven different RNA extraction methods: manual methods (TRIzol™), semi-automatic methods (QIAGEN™, Bio-Rad, Monarch®, and Canvax™), and fully automatic methods (QIAcube™ and Maxwell®). The results showed significant differences in RNA quality and functionality depending on the RNA extraction and the matrix used. QIAcube™ and semi-automatic extraction methods were considered the best choices due to their low variability, good functionality, and low cost (P < 0.001). These data aided researchers or research service platforms (biobanks) in decision-making practices and emphasized the relevance of selecting RNA extraction methods in each experimental procedure or traceability study to ensure quality standards and reproducibility.
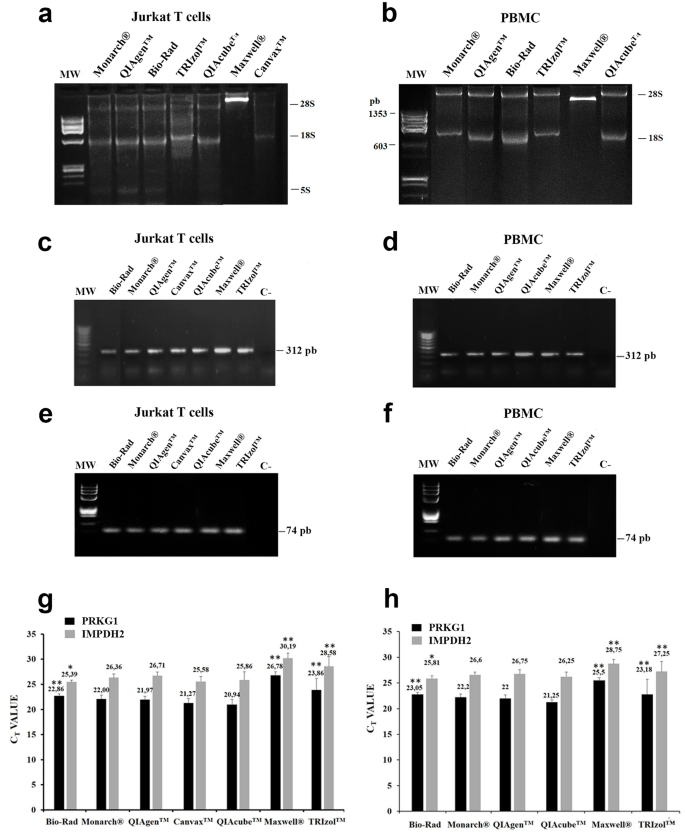
Figure 1. RNA Analysis in Human Jurkat T Cells and PBMCs [1]
2. Replacing the Detection of SARS-CoV-2 RNA with a New Mass Spectrometry-Based Detection Method
Accurate detection of SARS-CoV-2 RNA is crucial for stopping the spread of SARS-CoV-2. Studies aimed to evaluate the performance of the newly launched MassARRAY® SARS-CoV-2 Panel and compared it with the cobas® SARS-CoV-2 Test. The MassARRAY® SARS-CoV-2 Panel consists of five assays targeting different sequences of the SARS-CoV-2 genome. In clinical evaluation, 101 residual clinical samples were analyzed, and the results were compared. The samples had been tested for SARS-CoV-2 RNA using the cobas® SARS-CoV-2 Test. When testing accuracy with the MassARRAY® SARS-CoV-2 Panel, 25 out of 27 samples (92.6%) had correct results. When analyzing clinical samples with the MassARRAY® SARS-CoV-2 Panel and comparing it with the cobas® SARS-CoV-2 Test, results were consistent for 100 samples. One sample showed inconsistent results with the MassARRAY® SARS-CoV-2 detection team. When time-to-results were compared, the new assay showed longer total and hands-on times. The MassARRAY® SARS-CoV-2 panel exhibited good performance, proving suitable for routine diagnostic laboratories. Especially during phases of shortage of reagents and/or disposables, the new test system appeared as benefificial alternative to standard assays used for detection of SARS-CoV-2 RNA.
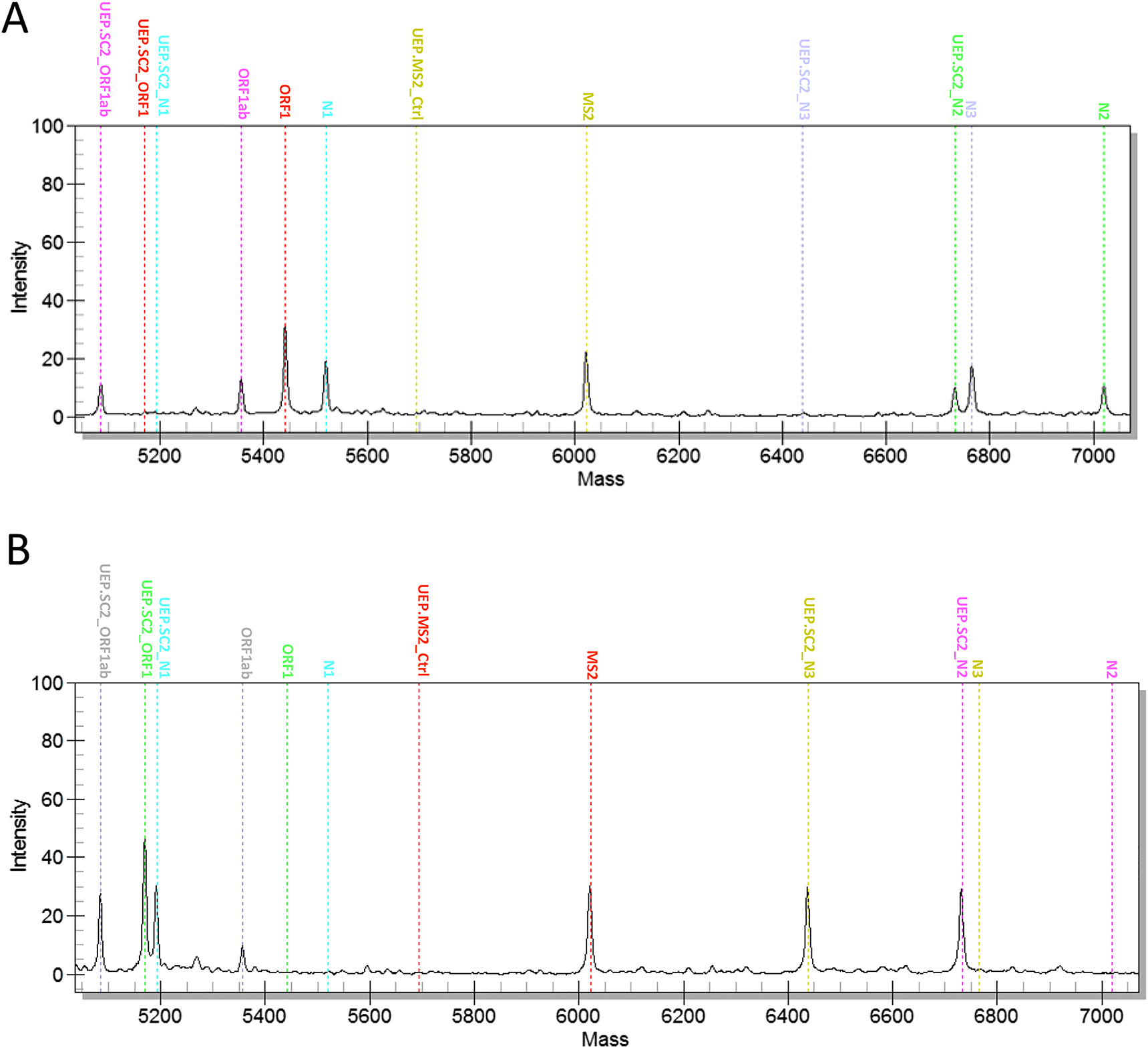
Figure 2. Example of Positive and Negative Results Obtained with the MassARRAY® SARS-CoV-2 Panel [2]
3. Ultra-Sensitive Detection of Bacillus Anthracis by Real-Time PCR Targeting Multicopy 16S rRNA Genes and Their Transcripts
Bacillus anthracis, a pathogen causing anthrax, poses a significant threat to human health. Due to the close genetic relationship between Bacillus anthracis and Bacillus cereus group species, identifying Bacillus anthracis is challenging. Therefore, molecular detection is based on species-specific PCR targeting single-copy genes. Studies have validated previously recognized multicopy targets, specifically single nucleotide polymorphism (SNP) present in 2-5 copies in each B.anthracis genome analyzed. To this end, a hydrolysis probe-based real-time PCR assay was developed and rigorously tested. This method was specific, as only B. anthracis DNA can produce a positive result, showing linear above 9 log10 units, high sensitivity with a limit of detection (LoD) of 2.9 copies per reaction. Although its LoD was not lower than that of existing single-copy PCR targets (dhp61 or PL3), the copy number of B. anthracis-specific 16S rRNA gene alleles was higher, thus providing a threshold (Ct) value ≤2 units. To further improve the detection limit, this method also was applied to reverse transcription PCR on16S rRNA transcripts. This PT-PCR research also exhibited linearity over 9 log10 units, high sensitivity with an LoD of 6.3 copies per reaction. In a series of dilution experiments, the sensitivity of the 16S RT-PCR method was found to be a thousand times higher than that of DNA-targeting methods. For molecular diagnostics, it is advisable to use the real-time RT-PCR assay variant, where both DNA and RNA can serve as templates (thus, no need for DNase treatment). If no RNA is present, this method could at least provide results equivalent to DNA-based methods, but it exceled even at the lowest residual rRNA concentrations.
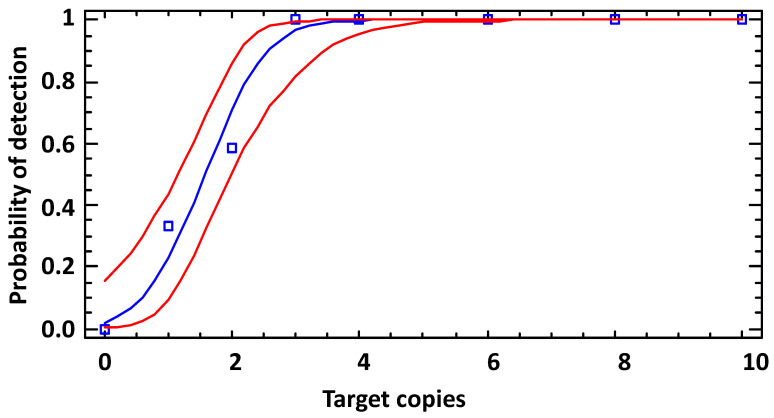
Figure 3. LoD of 16S rRNA SNP-PCR (Analytical Sensitivity) [3]
Sample Submission Requirements
1. Avoid Contamination with Impurities as Much as Possible
Services at MtoZ Biolabs
1. Complete Experimental Protocol
2. Related Instrument Parameters
3. Raw Experimental Data
4. Data Analysis Report
References
[1] Ortega-Pinazo J, Pacheco-Rodríguez MJ, Serrano-Castro PJ, Martínez B, Pinto-Medel MJ, Gómez-Zumaquero JM, Lago-Sampedro A, García-Díaz B, Estivill-Torrús G, Emilio Ferro Gallego P. Comparing RNA extraction methods to face the variations in RNA quality using two human biological matrices. Mol Biol Rep. 2023 Nov;50(11):9263-9271. doi: 10.1007/s11033-023-08761-2. Epub 2023 Oct 9. PMID: 37812354.
[2] Stelzl E, Kessler HH, Mustafa HG, Mustafa ME, Santner BI, Seier J, La Torre M, Haushofer AC. Alternative detection of SARS-CoV-2 RNA by a new assay based on mass spectrometry. Clin Chem Lab Med. 2021 Aug 12;59(12):1998-2002. doi: 10.1515/cclm-2021-0483. PMID: 34388325.
[3] Braun P, Nguyen MD, Walter MC, Grass G. Ultrasensitive Detection of Bacillus anthracis by Real-Time PCR Targeting a Polymorphism in Multi-Copy 16S rRNA Genes and Their Transcripts. Int J Mol Sci. 2021 Nov 12;22(22):12224. doi: 10.3390/ijms222212224. PMID: 34830105; PMCID: PMC8618755.
How to order?







