Fungus-derived Exosome Research and Applications
Fungus-derived Exosome Research and Applications focuses on exosomes secreted by fungal cells, encompassing foundational research such as extraction and purification, omics analysis, and functional validation, as well as application development in areas including disease diagnostics, vaccine development, immune modulation, drug delivery, and quorum sensing regulation.
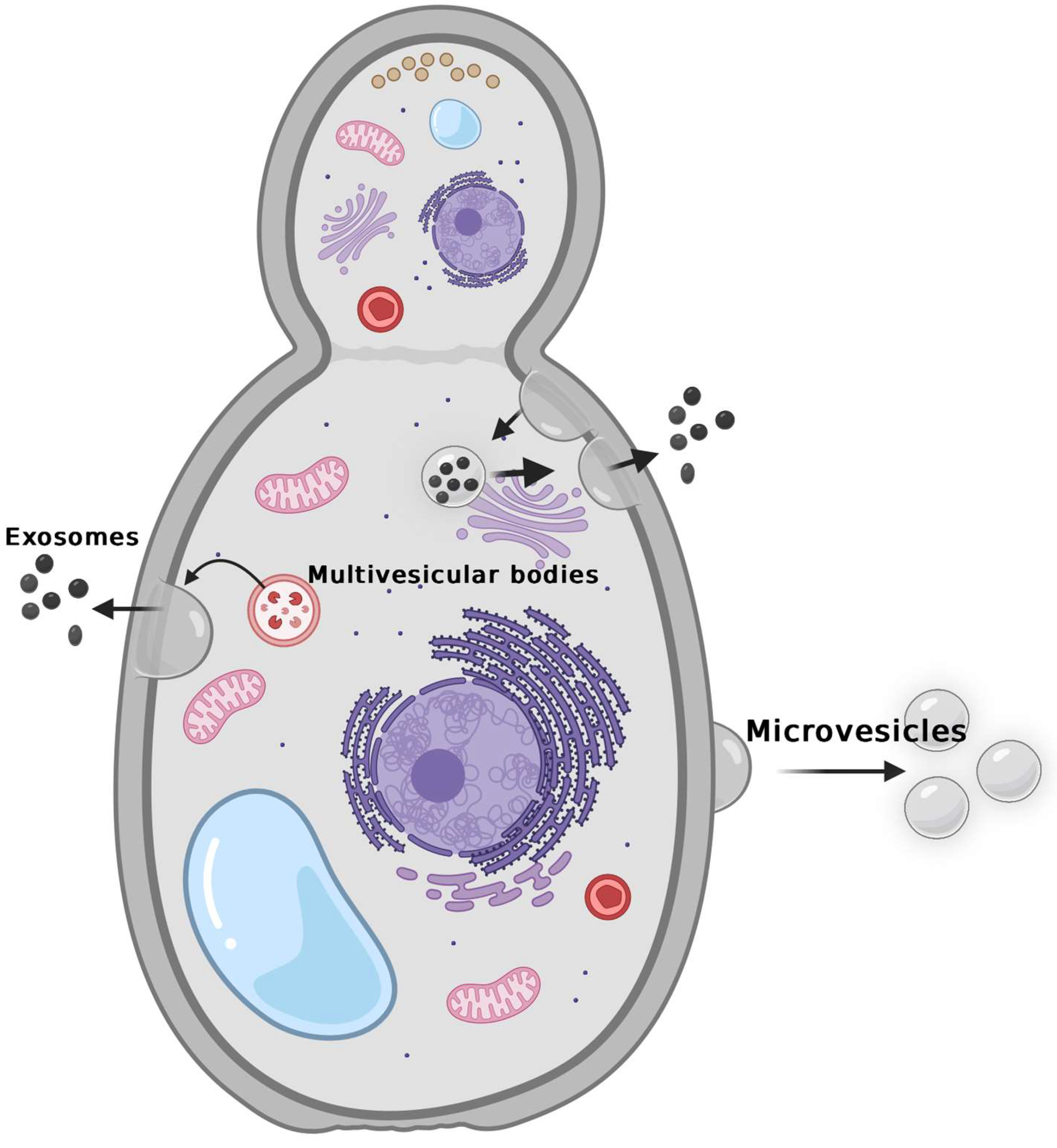
Ikeda MAK. et al. Microbiology Research. 2024.
Fungus-derived exosomes are small vesicles generated by fungal cells through intracellular multivesicular body (MVB) pathways or plasma membrane budding. These vesicles are rich in bioactive molecules such as proteins, lipids, polysaccharides, RNAs, and virulence factors. Fungus-derived exosomes play key regulatory roles in quorum sensing, morphological modulation, signal transduction, and infection establishment, and have recently been recognized as critical mediators of fungus-host interactions. Their bioactive contents can interact with host cell surface receptors or be internalized to modulate host transcriptomic profiles and immune responses. Unlike mammalian exosomes, fungal exosomes must traverse a rigid cell wall, which results in unique structural and biogenetic characteristics. This distinctiveness not only expands the frontier of exosome biology but also offers new perspectives for antifungal strategies, biomarker discovery, and nano-delivery system development.
MtoZ Biolabs provides comprehensive Fungus-derived Exosome Research and Applications service including systematic fungal exosome extraction, purification, identification, and compositional analysis. Our services support both basic and translational research, enabling clients to explore the molecular composition, biological functions, and application potential of fungal secreted vesicles. These include studies on fungal pathogenicity, host immune modulation, biomarker discovery, and natural product delivery. Leveraging a robust platform for exosome isolation, omics profiling, and functional validation, MtoZ Biolabs supports research across model fungi, pathogenic fungi, and industrial fungal strains.
Analysis Workflow
MtoZ Biolabs offers a comprehensive workflow for fungus-derived exosome research, covering the following core steps:
1. Fungal Cultivation
Multiple culture conditions (liquid/solid media) and growth phase control strategies are provided to ensure spatiotemporal consistency in exosome production.
2. Exosome Extraction and Purification
Classic methods such as ultracentrifugation, density gradient centrifugation, and size-exclusion chromatography (SEC) are used to isolate high-purity exosomes.
3. Morphology and Particle Size Analysis
Transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and dynamic light scattering (DLS) are employed to confirm particle size distribution and morphological features.
4. Compositional Analysis
Proteomics, transcriptomics, metabolomics, and other omics platforms are utilized to comprehensively profile the functional molecules carried by fungal exosomes.
Applications
MtoZ Biolabs' Fungus-derived Exosome Research and Applications service supports a broad range of research and development directions:
Fungal Pathogenic Mechanism Research
Explore how fungi utilize vesicles to mediate immune evasion, inflammatory activation, and tissue damage.
Antifungal Target Discovery
Identify novel virulence factors or efflux system components through vesicle proteomics.
Vaccine or Diagnostic Biomarker Development
Screen for surface antigens or RNA content within vesicles as potential biomarkers.
Natural Product Delivery Research
Use engineered vesicles to deliver antimicrobial peptides, polysaccharides, or plant-derived compounds.
Biocontrol and Agricultural Applications
Investigate vesicle-mediated virulence or resistance signaling in plant fungal diseases to support eco-friendly crop protection strategies.
Quorum Sensing and Biofilm Regulation
Study the role of vesicles in yeast-to-hyphae transition, biofilm formation, and fungal communication networks.
Service Advantages
1. Efficient Isolation: Integrated multi-platform exosome purification ensures high yield and purity.
2. Multi-Dimensional Analysis: Supports structural, compositional, and functional analysis for both mechanistic studies and product development.
3. Customized Service: Experimental designs tailored to fungal species and research objectives.
4. High-Throughput Omics Platforms: Enables integrative proteomics, transcriptomics, and lipidomics analyses.
5. Strict Quality Control: Each batch is accompanied by a full QC report to ensure data integrity and reproducibility.
FAQ
Q. Are there Significant Extraction Differences between Yeast and Filamentous Fungi-Derived Exosomes?
Yes. Yeast and filamentous fungi differ significantly in cell wall structure, growth behavior, and vesicle secretion levels. Filamentous fungi often require stronger mechanical or enzymatic pretreatment for efficient vesicle release, while yeast tends to secrete more readily collectable exosomes. MtoZ Biolabs provides strain-specific extraction protocols optimized for culture medium, growth stage, supernatant processing, and centrifugation conditions to ensure high-quality exosome recovery.
Q. Do you Support Deep Omics Analyses such as Proteomics, Lipidomics, and RNA Profiling (Especially sRNA)?
Absolutely. We offer in-depth multi-omics profiling of exosomes, including LC-MS/MS-based proteomics, shotgun lipidomics, and small RNA-seq/total RNA-seq. For fungal species, we integrate fungi-specific databases (e.g., FungiDB, JGI MycoCosm) and perform GO/KEGG annotation. Customized analyses such as differential expression, pathway enrichment, and network visualization are available to support functional and regulatory insights into fungal exosomes.
Case Study
This study conducted a systematic analysis of extracellular vesicles (EVs) from Candida albicans to explore their role in fungal self-regulation. Transcriptomic profiling (RNA-seq) revealed that EV-treated cells exhibited altered gene expression in the L-arginine/NO pathway. By combining nitric oxide (NO) and reactive oxygen species (ROS) measurements, apoptosis assays, and host cell damage analyses, the researchers demonstrated that C. albicans EVs promoted fungal growth and enhanced pathogenicity by activating NO synthesis, reducing ROS accumulation, and suppressing apoptosis. This work highlights the critical role of fungal-derived vesicles in regulating metabolism, immune evasion, and virulence.
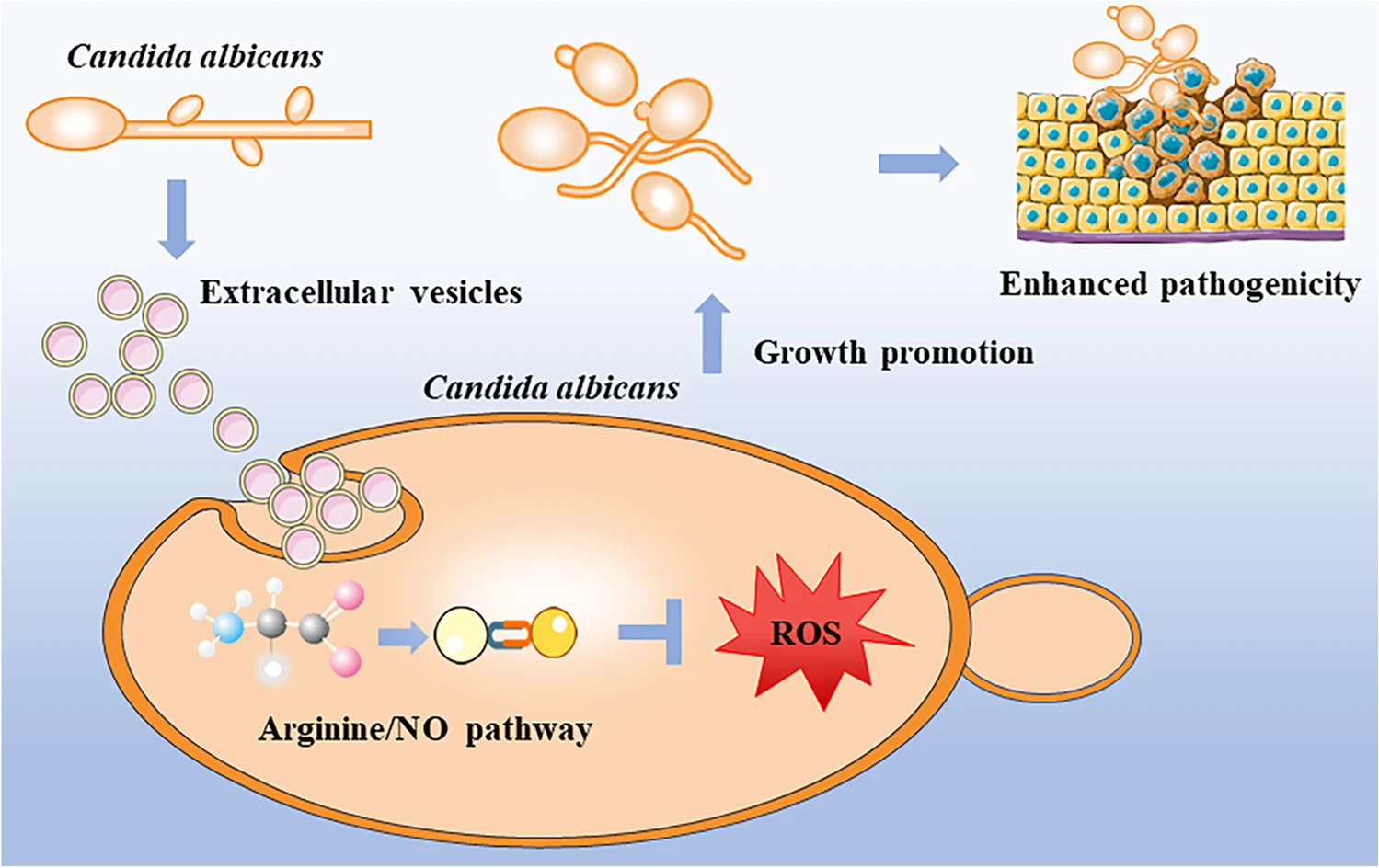
Wei Y. et al. Appl Microbiol Biotechnol. 2023.
How to order?







