Functional Proteomics Service
Functional proteomics is a crucial branch of proteomics dedicated to studying protein functions, interactions, and their dynamic changes within biological systems. With the rapid advancement of genomics and proteomics technologies, functional proteomics plays an irreplaceable role in elucidating fundamental mechanisms of life, uncovering the molecular basis of disease progression, and guiding novel drug development.
Functional proteomics has evolved from traditional proteomics with a stronger emphasis on investigating the functional properties of proteins and their dynamic alterations within cells. Functional proteomics service can explore the mechanisms by which proteins contribute to cellular signal transduction, metabolic regulation, cell cycle progression, and immune responses, thereby providing new insights and strategies for disease diagnosis and treatment.
Functional proteomics service has broad applications across various fields. For instance, in cancer research, functional proteomics technologies have enabled the identification of numerous protein biomarkers associated with tumor initiation, progression, and metastasis. These biomarkers not only facilitate early cancer detection but also serve as a foundation for personalized treatment approaches. In neuroscience, functional proteomics is employed to dissect the molecular mechanisms underlying brain development, neuronal function, and neurological disorders, offering new therapeutic strategies for neurodegenerative diseases.
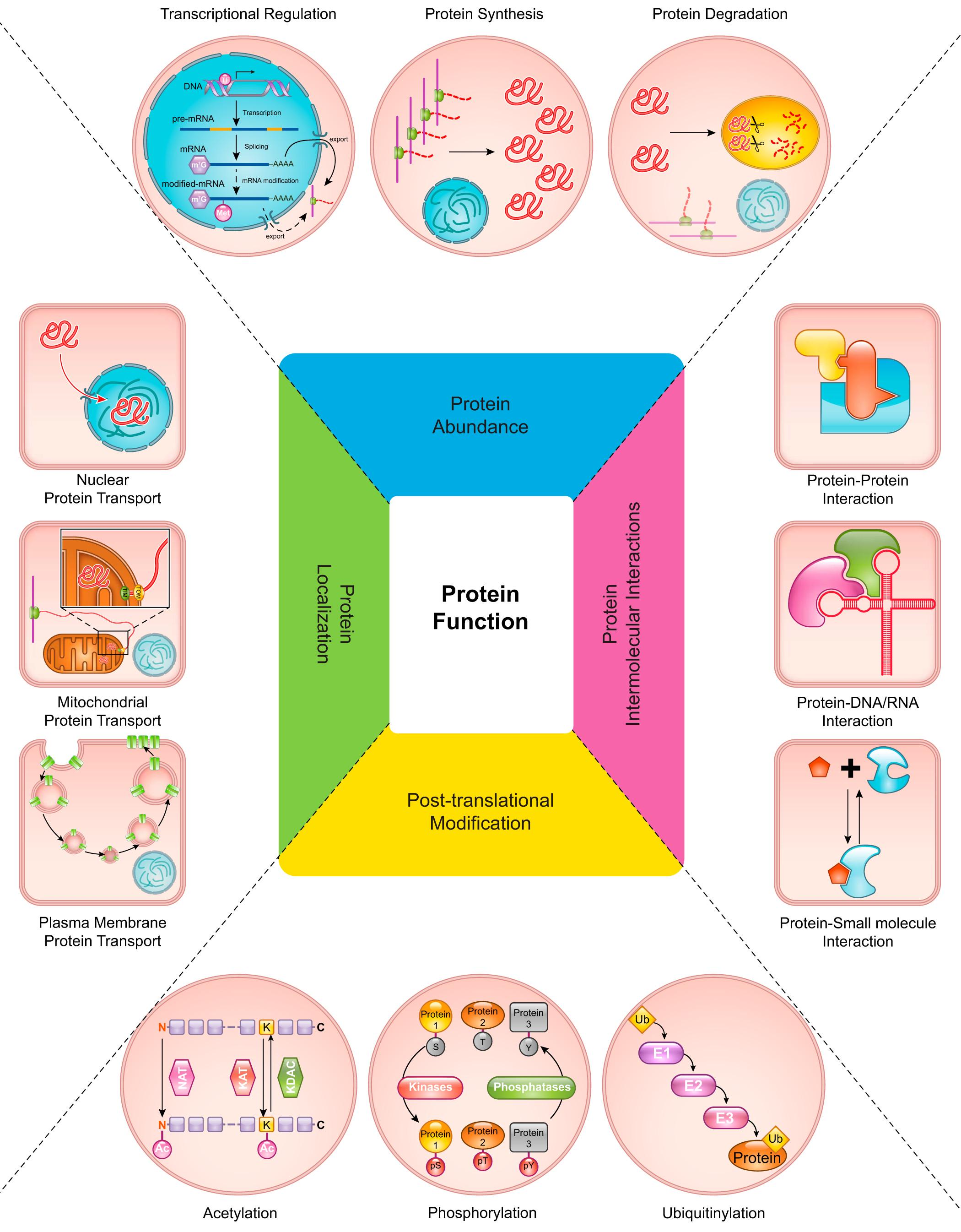
Rinschen, M M. et al. Physiological Reviews, 2018.
Figure 1. Overview of Functional Proteomic Regulatory Mechanisms.
Services at MtoZ Biolabs
Leveraging advanced analytical instruments, the comprehensive functional proteomics service provided by MtoZ Biolabs, including but not limited to:
1. Quantitative Proteomics
MtoZ Biolabs offers multiple quantitative proteomics techniques, including but not limited to:
(1) Label-Free Quantitative Proteomics: A mass spectrometry-based label-free quantitative proteomics method for qualitative and relative quantification of proteins in samples. Compared to traditional labeling techniques, the label-free approach is more straightforward and flexible as it does not require chemical labeling of protein samples.
(2) DIA Quantitative Proteomics: Data-Independent Acquisition (DIA) technology segments the entire mass spectrometry scanning range into multiple small windows and sequentially fragments all ions within each window. This method enables the quantification of nearly all detectable molecules in complex samples, overcoming the limitations of the DDA approach where low-abundance proteins may be undetectable due to suppression.
(3) Low-Abundance Proteomics: This technique integrates specialized sample preparation methods with minimal peptide/protein loss and high-sensitivity mass spectrometry analysis to ensure normal data acquisition from low-abundance samples.
(4) 4D Quantitative Proteomics: In addition to conventional mass spectrometry data (retention time, mass-to-charge ratio, and ion intensity), 4D proteomics incorporates collisional cross-section (CCS) measurements, improving identification depth and sensitivity.
(5) TMT Quantitative Proteomics: A mass spectrometry-based multiplexed labeling technique suitable for large-scale protein quantification analysis.
2. Mass Spectrometry-Based Post-Translational Modification (PTM) Proteomics
MtoZ Biolabs offers a variety of post-translational modification (PTM) analysis services, including but not limited to:
(1) Phosphoproteomics Quantification: Common modification sites include Ser/Thr/Tyr. Relevant applications include signal transduction, cell cycle regulation, regulatory mechanisms, stress resistance, growth and development, and cancer mechanisms.
(2) Disulfide Bond Proteomics: Common modification site is Cys. Relevant applications include protein structural stability, protein activity, biopharmaceutical design, drug efficacy, and safety evaluation.
(3) SUMOylation Quantification Proteomics: Common modification site is Lys. Relevant applications include embryonic development, cardiac and craniofacial development, plant immune responses, DNA damage modifications, and tumor drug resistance.
(4) Methylation Quantification Proteomics: Common modification sites include Arg/Lys. Relevant applications include epigenetics, cancer mechanisms, aging, neurodegenerative diseases, protein translocation and signal transduction, and histone functionality.
(5) Acetylation Quantification Proteomics: Common modification site is Lys. Relevant applications include gene expression regulation, cellular defense mechanisms, apoptosis and metabolism, cell cycle, transcription activation and silencing, protein stability, and neurodegenerative disorders.
(6) Glycosylation Quantification Proteomics: Common modification sites include Asn (N-linked) and Ser/Thr (O-linked). Relevant applications include cell recognition, differentiation, stress response, signal transduction, immune response, neurodegenerative diseases, metabolic disorders, and infectious disease research.
(7) Lactylation Quantification Proteomics: Common modification site is Lys. Relevant applications include glycolysis-related cellular functions, macrophage polarization, nervous system regulation, tumor development, sepsis, immune disease progression, and rice grain development.
(8) Ubiquitination Quantification Proteomics: Common modification site is Lys. Relevant applications include cell cycle, apoptosis, protein degradation, defense mechanisms, photomorphogenesis, signal transduction, plant growth and development, cancer, and neurodegenerative diseases.
3. Chemical Proteomics
MtoZ Biolabs also provides chemical proteomics services, including but not limited to:
(1) Activity-Based Protein Profiling: Used to study protein activity and function, particularly in drug development and disease mechanism research.
Service Advantages
1. Advanced Analytical Instruments
MtoZ Biolabs is equipped with high-resolution mass spectrometers, including the Thermo Fisher Orbitrap Fusion Lumos platform, combined with Nano-LC, to establish a functional proteomics service platform that delivers high-sensitivity and high-resolution protein analysis services.
2. Diverse Analytical Techniques
We offer a variety of proteomics quantification methods tailored to your samples, including Label-free, iTRAQ, TMT, SILAC, 2D-DIGE, DIA, SWATH, relative quantification, semi-quantification, and absolute quantification, meeting a wide range of research needs.
3. One-Stop Service
MtoZ Biolabs provides end-to-end services, from sample preparation to data analysis, ensuring that clients receive high-quality and efficient proteomics research outcomes. With experienced technical personnel and a well-established quality control workflow, we handle various sample types and customize projects based on specific research needs.
4. Transparent Pricing
Our pricing structure is transparent, with no hidden or additional fees.
5. High Data Quality
Our deep data coverage and strict quality control measures guarantee comprehensive and reliable data reports for our clients.
6. High-Throughput Capabilities
We are capable of processing large volumes of samples efficiently while maintaining high-quality standards and quick turnaround times.
7. Customized Services
We provide tailored analysis solutions based on client-specific requirements, ensuring optimal results for each research project.
Applications
1. Disease Diagnosis
(1) Biomarkers: Through high-throughput screening and identification of disease-specific biomarker molecules, critical evidence can be provided for disease diagnosis, particularly for early-stage detection. For example, prostate-specific antigen (PSA) plays a crucial role in the diagnosis of prostate cancer.
(2) Monitoring Disease Progression: Analysis of protein composition in patients' blood through functional proteomics service can monitor disease progression and therapeutic effects. For instance, proteomics techniques can detect protein variations associated with tumors, cardiovascular diseases, and neurodegenerative disorders.
2. Drug Development
(1) Identification of Drug Targets: Proteomics methods are used to study the interactions between proteins in healthy and diseased organisms, as well as the pathological basis of diseases, to identify potential drug targets.
(2) Investigation of Drug Mechanisms: By analyzing changes in intracellular proteins after drug treatment, researchers can gain a deeper understanding of the mechanism of action and potential side effects of drugs. For example, in studies on the anticancer effects of taxane-based drugs, Bauer et al. conducted a proteomics analysis on breast cancer patients who experienced recurrence after taxane treatment and discovered that α-defensin could serve as a biomarker for predicting the therapeutic efficacy of this class of drugs in breast cancer treatment.
3. Plant Stress Resistance Research
(1) Environmental Signal Response: Functional proteomics service can be used to study changes in protein expression in plants under different environmental conditions, thereby uncovering plant stress resistance mechanisms. For example, in a study on maize subjected to forced hypoxia and low oxygen conditions, Chang et al. identified 46 related proteins through mass spectrometry, revealing that the effects of low oxygen treatment were not only induced by reduced oxygen levels but also led to an increase in glycolytic enzymes.
(2) Crop Improvement: Proteomics technology enables the identification of proteins associated with plant stress resistance, providing a theoretical basis for crop improvement. For instance, studying the protein expression changes in crops such as cotton, potatoes, and rice under different environmental conditions can help develop more stress-resistant varieties.
4. Pathogenic Microorganism Research
(1) Pathogenic Mechanisms: Proteomics technology can be utilized to study the protein expression changes in pathogenic microorganisms, thereby revealing their pathogenic mechanisms. For example, Fernandez et al. employed two-dimensional gel electrophoresis and MALDI-TOF MS technology to analyze protein spots isolated from Botrytis cinerea cultured in carboxymethyl cellulose. Their findings identified numerous proteins that contribute to the pathogen's virulence.
(2) Vaccine Development: Functional proteomics service can be used to identify specific biomarkers of pathogenic microorganisms, providing new insights for vaccine development. For instance, by analyzing the protein expression changes of pathogenic microorganisms at different growth stages, potential vaccine candidate proteins can be identified.
Case Study
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest cancers, with a five-year survival rate below 10%. Developing reliable early diagnostic biomarkers and effective targeted therapies is critical. The tumor microenvironment (TME) is a hallmark of PDAC, consisting of abundant non-malignant stromal cells and extracellular matrix components, which interact with tumor cells to promote proliferation, metastasis, and drug resistance. Mass spectrometry-based proteomics enables systematic exploration of intercellular signal transduction in PDAC. However, traditional proteomics research using bulk tumor samples or cell cultures often yields averaged data, making it difficult to reveal precise functional proteomic characteristics in clinical samples. In order to solve this problem, researchers developed the clinical functional protein omics analysis strategy TMEPro, which started with the underlying principle, wet experiment and dry experiment, and used a variety of chemometrics and chemical biology strategies to analyze the characteristics of functional protein groups in tumor samples in multiple dimensions, thus systematically solving the above technical problems.
By developing TMEPro, researchers conducted a direct spatiotemporal proteomic analysis of minute PDAC clinical samples from five different dimensions, overcoming the limitations of traditional research methods that rely on bulk tissue samples or cultured cell lines. Through further integration of multidimensional bioinformatics analysis, they constructed a comprehensive intercellular signaling network map, revealing functional secreted ligand-receptor membrane protein interactions between cancer cells and stromal cells. This study identified extracellular domain cleavage of receptor tyrosine kinase AXL in cancer cells, as well as an interactive signaling axis between cancer cells and stromal cells. These findings provide a valuable functional proteomics database and potential molecular classification and targeted therapeutic strategies for the precision diagnosis and treatment of pancreatic cancer.
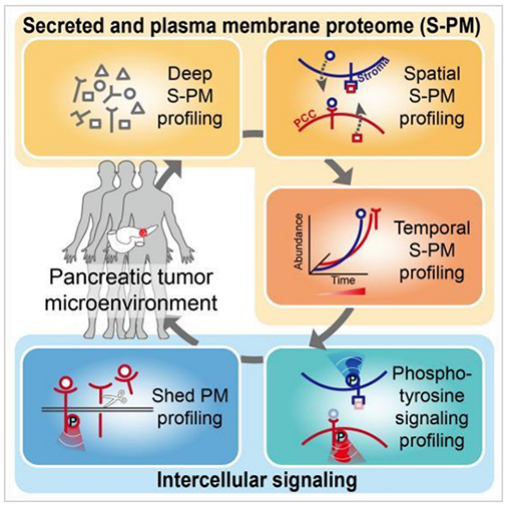
Huang, P. et al. Nature, 2024.
Figure 2. Schematic Diagram of the TMEPro-Analyzed Intercellular Signaling Network in the Pancreatic Cancer Tumor Microenvironment.
Functional proteomics, as a vital branch of proteomics, plays an irreplaceable role in deciphering fundamental biological mechanisms, elucidating the molecular basis of disease progression, and guiding new drug development. MtoZ Biolabs leverages cutting-edge mass spectrometry and imaging technologies to deeply investigate protein functions and their dynamic changes within cells, offering novel insights and methodologies for disease diagnosis and treatment.
As a professional provider of chromatography and mass spectrometry services, MtoZ Biolabs has made significant contributions in the functional proteomics service field. The company employs advanced mass spectrometry and imaging technologies to deliver precise and reliable functional proteomics services for researchers, thereby driving the in-depth development of functional proteomics research. In the future, with continuous technological advancements and expanding applications, functional proteomics will play an increasingly important role across various fields. We are committed to making greater contributions to human health.
Our ultimate goal is to provide faster, high-throughput, and cost-effective analyses with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







