Exosomal Whole Transcriptome Sequencing Service
Exosomes are small extracellular vesicles released by most cell types into various biological fluids such as blood, urine, saliva, and cerebrospinal fluid. These vesicles carry a wide array of RNA species, including messenger RNA (mRNA), long non-coding RNA (lncRNA), circular RNA (circRNA), and small RNAs, which reflect the physiological and pathological status of their originating cells. As exosomal RNA is protected from degradation by the vesicle’s lipid bilayer, it represents a stable and accessible source of transcriptomic information. Whole transcriptome sequencing of exosomal RNA has emerged as a powerful method to comprehensively capture the coding and non-coding transcriptomic landscape of exosomes. This approach enables the discovery of novel biomarkers, investigation of gene expression changes, and exploration of regulatory RNA networks associated with disease development, progression, and therapeutic response. To support researchers in decoding exosomal RNA profiles, MtoZ Biolabs offers a specialized Exosomal Whole Transcriptome Sequencing Service that provides high-resolution insights into both coding and non-coding RNA content using advanced next-generation sequencing (NGS) platforms and optimized RNA preparation protocols.
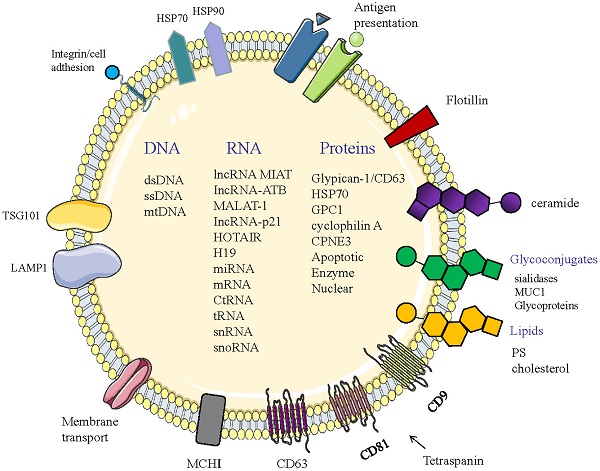
Figure 1. Composition of Exosomes
Services at MtoZ Biolabs
MtoZ Biolabs offers a comprehensive suite of analytical solutions as part of our Exosomal Whole Transcriptome Sequencing Service, enabling researchers to extract meaningful biological insights from exosomal RNA. Our Exosomal Whole Transcriptome Sequencing Services are designed to support diverse research goals, from transcript discovery to expression quantification and structural variant analysis.
1. Transcript Diversity Profiling: Comprehensive detection of mRNAs, lncRNAs, circRNAs, and splicing isoforms to capture exosomal transcript complexity.
2. Quantitative Gene Expression Analysis: Accurate measurement of gene expression levels with support for differential analysis across sample groups.
3. Novel Transcript Discovery: Identification of previously unannotated transcripts and novel genes through reference-guided and de novo assembly, revealing new molecular targets.
4. Wide Dynamic Range Detection: Sensitive detection of both high- and low-abundance transcripts, ensuring comprehensive transcriptome coverage even from minute exosomal RNA inputs.
5. Functional Insights and Pathway Annotation: GO, KEGG, and network enrichment analyses to uncover the biological roles and regulatory pathways of exosomal RNAs.
6. Structural Variant Detection: Detection of SNPs, insertions/deletions (indels), alternative splicing events, and transcript fusions within exosomal RNA to reveal structural and regulatory alterations.
7. Single-Cell Resolution (Optional): Integration with single-cell sequencing data to map exosomal RNA signatures to specific cell types or functional states.
Analysis Workflow
Our standardized workflow of Exosomal Whole Transcriptome Sequencing Service ensures reliable and reproducible results across a wide range of sample types:
1. Exosome Isolation
Exosomes are isolated from biofluids or cell culture media using ultracentrifugation, size-exclusion chromatography, immunoprecipitation, or microfluidic platforms, with strict quality control for particle size and purity.
2. RNA Extraction and Quality Control
Total RNA is extracted from exosomes. RNA integrity and concentration are assessed using advanced platforms.
3. Library Preparation
The RNA is then fragmented and reverse-transcribed into cDNA. Adapters are ligated, and libraries are PCR-amplified under optimized conditions to preserve transcript diversity and minimize bias.
4. Sequencing
Libraries are sequenced using Illumina platforms, generating high-quality reads with customizable sequencing depth based on project needs.
5. Bioinformatics Analysis
Data are processed through rigorous pipelines, including quality control, alignment, transcript quantification, differential expression analysis, and functional annotation. Optional modules include pathway enrichment, novel transcript discovery, and biomarker candidate screening.
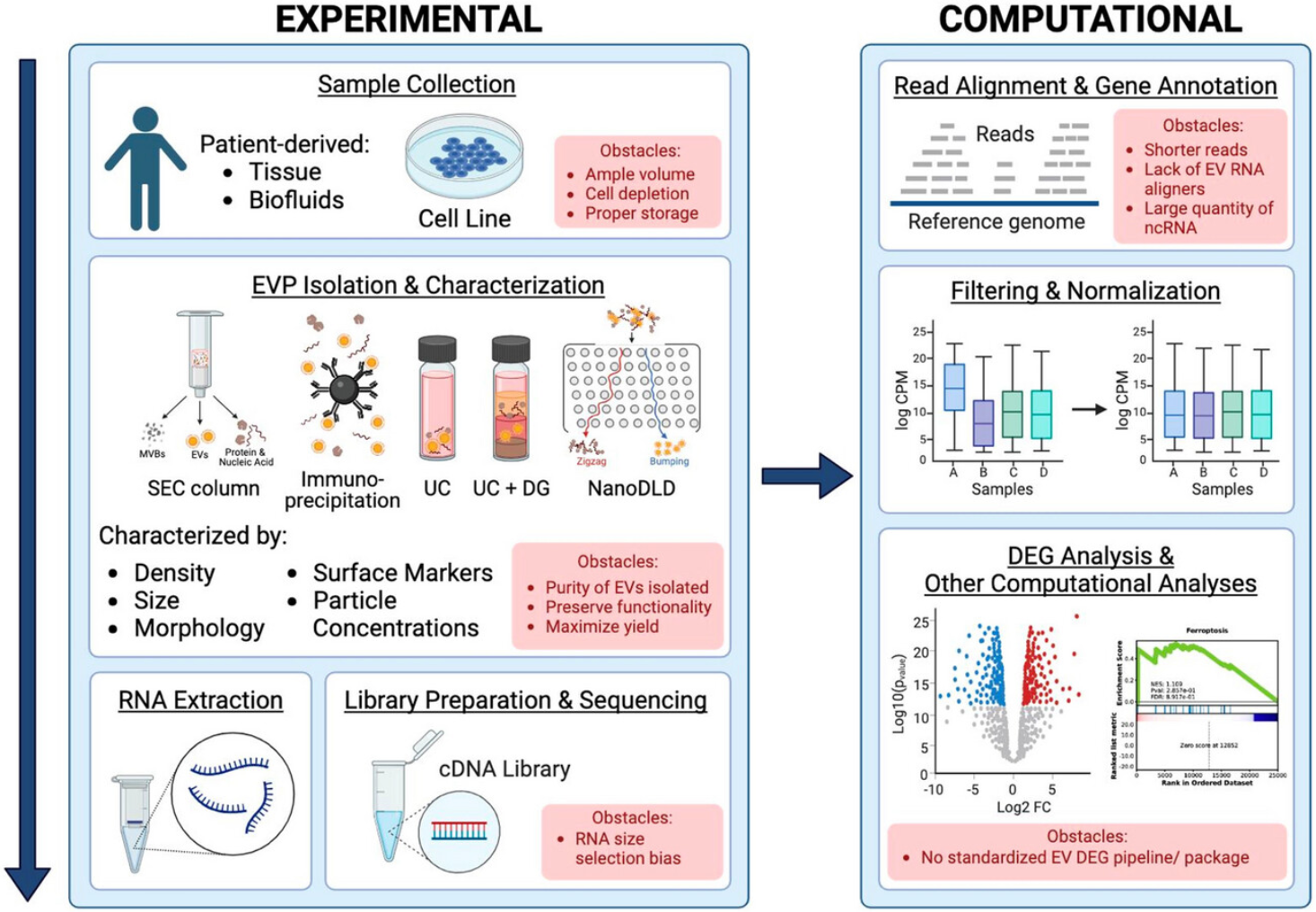
Figure 2. Workflow for Extracellular Vesicles Whole Transcriptomic Analysis
Service Advantages
☑️One-Stop Service: From exosome isolation to data interpretation, we provide an integrated service covering RNA extraction, library preparation, sequencing, and bioinformatics analysis—saving you time and ensuring consistency across all steps.
☑️Comprehensive RNA Profiling: Our platform captures a wide spectrum of RNA species, including mRNAs, lncRNAs, and circRNAs, enabling in-depth exploration of exosomal transcriptomes.
☑️Expert Bioinformatics Support: We deliver publication-ready analyses supported by experienced bioinformatics scientists.
☑️High Sensitivity and Data Quality: Using advanced sequencing platforms, we ensure sensitive detection of both high- and low-abundance RNAs with reliable data output.
☑️Flexible and Customizable Analysis: Our services can be tailored to your project needs, including options for single-cell integration, or biomarker validation.
Applications
1. Biomedical Research
Decode exosome-mediated gene regulation and intercellular communication to advance the understanding of biological processes such as cell signaling, differentiation, and homeostasis.
2. Molecular Diagnostics Development
Facilitate the discovery and validation of exosomal RNA biomarkers for non-invasive diagnostic assays, with potential applications in early detection, prognosis, and therapeutic monitoring.
3. Translational and Clinical Research
Bridge basic findings with clinical applications by profiling exosomal transcriptomes in patient-derived biofluids, enabling precision medicine studies and companion diagnostics development.
5. Drug Development
Monitor transcriptomic changes in exosomal RNA in response to pharmacological agents to assess drug efficacy, mechanism of action, and potential off-target effects.
Case Study
Exosomal tRFs Profiling in Colorectal Cancer
A recent study utilized exosomal whole transcriptome sequencing to investigate tRNA-derived fragments (tRFs) in colorectal cancer (CRC). Researchers profiled RNA from both tumor tissues and plasma-derived exosomes, identifying three upregulated tRFs (tRF-3022b, tRF-3030b, and tRF-5008b) in CRC patients. These tRFs were not only elevated in tumor tissues but also enriched in exosomes, suggesting active secretion by cancer cells. Functional assays revealed that silencing tRF-3022b suppressed CRC cell proliferation and induced apoptosis, while also promoting M2 macrophage polarization via interaction with LGALS1 and MIF. This study demonstrates the utility of exosomal whole transcriptome sequencing in uncovering non-coding RNA biomarkers and mechanistic targets in cancer progression and immune modulation.
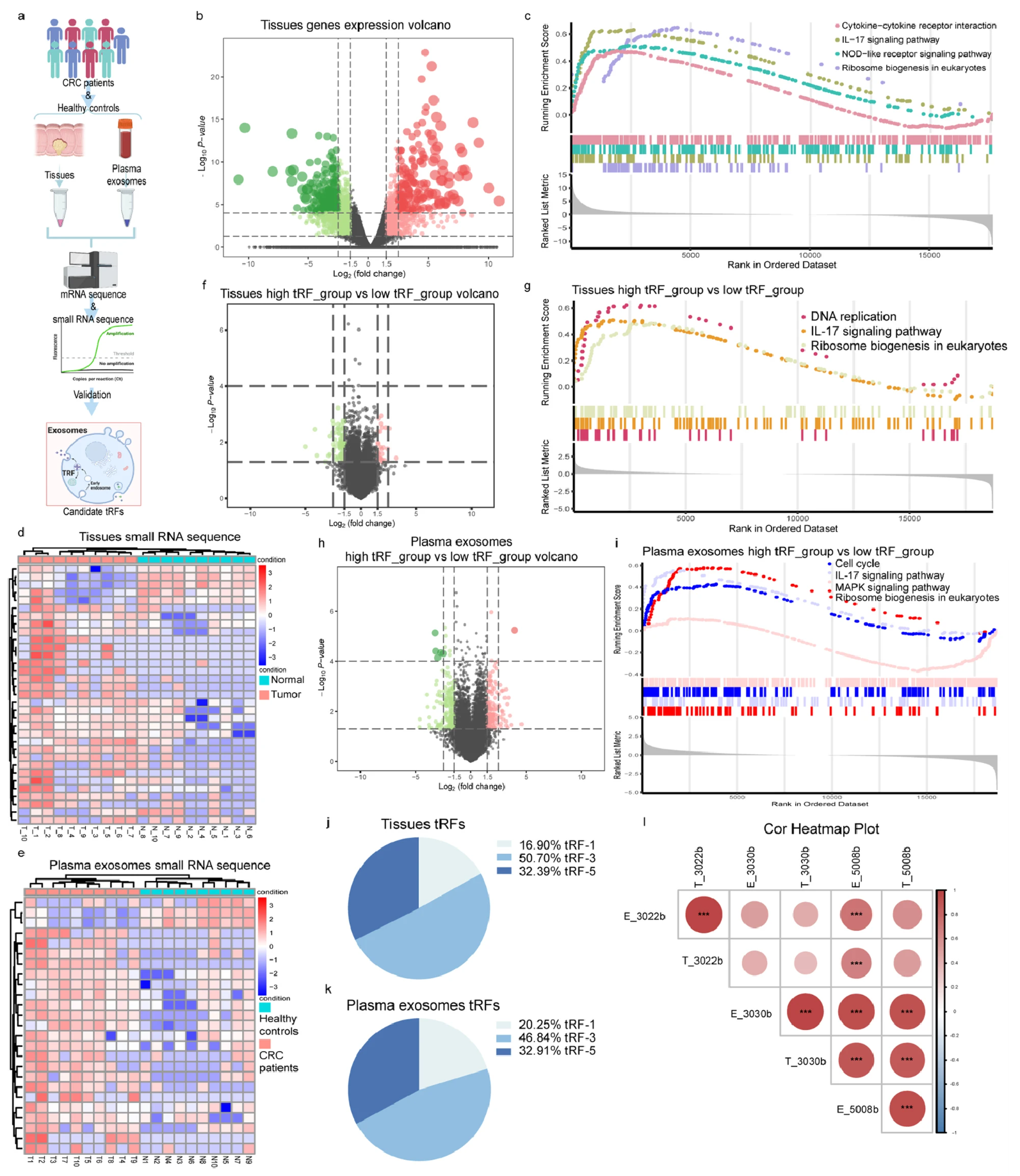
Figure 3. Screening and Profiling of tRFs in the Tissues and Plasma Exosomes from CRC Patients and Healthy Controls
To accelerate your exosome research with high-throughput transcriptomic insights, contact MtoZ Biolabs today. Our scientific team is ready to assist with experimental design, sample submission, and technical consultation to ensure your project success.
How to order?







