Exosomal cfDNA Isolation and Profiling Service
Exosomal cfDNA Isolation and Profiling Service extracts cell-free DNA (cfDNA) from exosomes and performs sequence analysis, methylation analysis, or fragment analysis to study its role in disease diagnosis, tumor monitoring, and microenvironment regulation. Cell-free DNA (cfDNA) refers to DNA fragments that circulate freely in bodily fluids such as blood, urine, and saliva, typically originating from cell apoptosis, necrosis, or active secretion. In recent years, cfDNA has demonstrated broad potential as a non-invasive biomarker in liquid biopsies, cancer detection, and disease monitoring. However, cfDNA is not only present in plasma or serum but can also be encapsulated by exosomes and stably transported to distant cells, enhancing its stability in bodily fluids and protecting it from degradation by nucleases. Compared with traditional plasma cfDNA analysis, exosomal cfDNA, thanks to its membrane protection, exhibits higher stability and can provide cell-of-origin-specific information. Based on efficient exosomal cfDNA extraction techniques and various DNA analysis technologies, MtoZ Biolabs offers the Exosomal cfDNA Isolation and Profiling Service to enable sequencing and quantitative analysis of exosomal cfDNA, thereby investigating its role in disease diagnosis, tumor monitoring, and microenvironment regulation.
Analysis Workflow
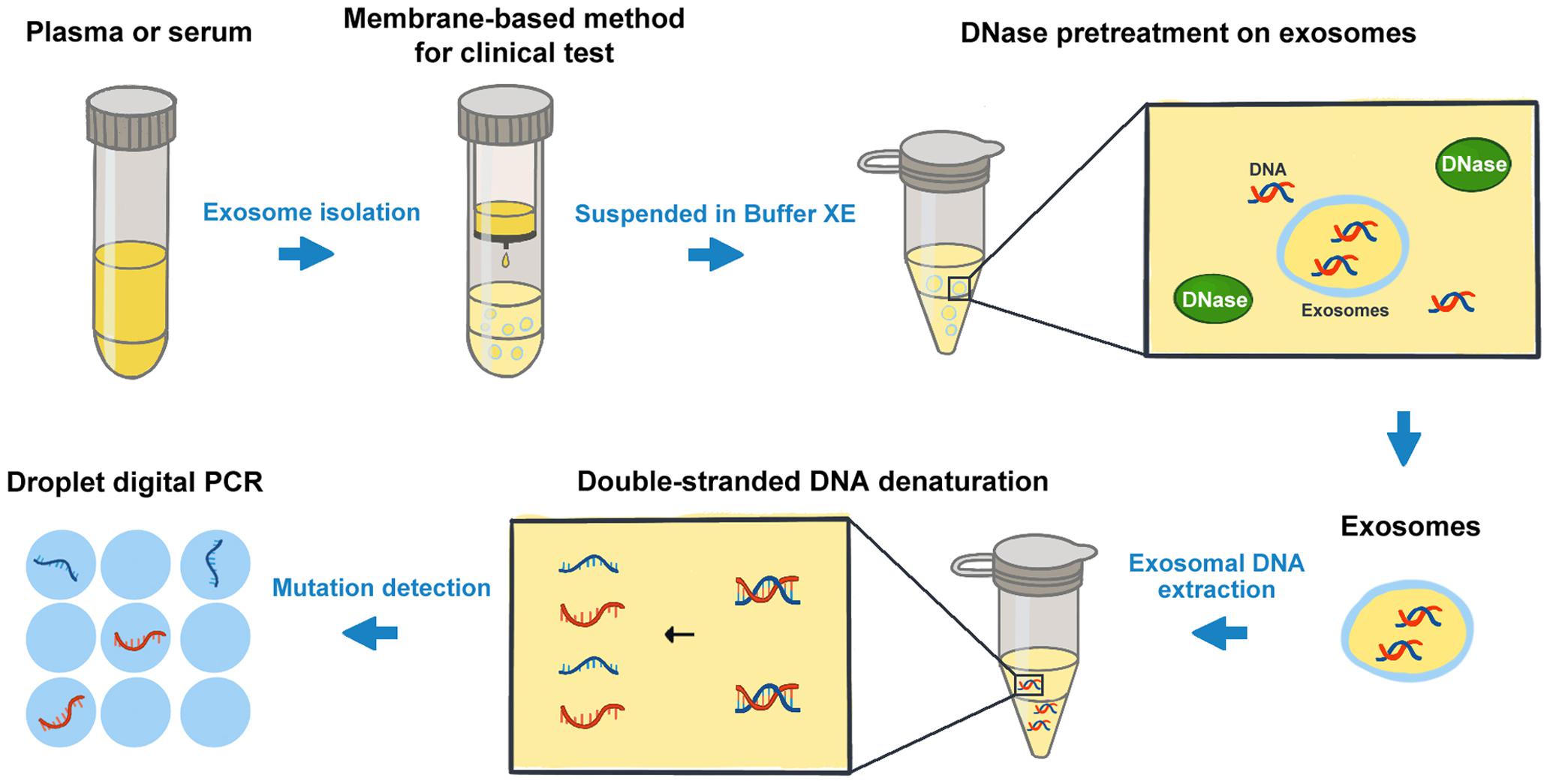
Wang Z Y. et al. Frontiers in Oncology. 2020.
1. Exosome Isolation
Choose an appropriate method (ultracentrifugation, SEC, immunoaffinity capture, etc.) to extract exosomes.
2. cfDNA Extraction
Lyse the exosomes and extract the cfDNA.
3. cfDNA Quantification and Quality Control
Measure the DNA concentration and fragment size.
4. cfDNA Analysis
Depending on research requirements, choose techniques such as NGS, qPCR, or digital PCR for cfDNA analysis.
Applications
Application examples of Exosomal cfDNA Isolation and Profiling Service:
Cancer Detection and Monitoring
By detecting tumor-specific mutations in plasma exosomal cfDNA, biomarkers for non-invasive early cancer detection and dynamic monitoring can be identified.
Non-Invasive Disease Diagnosis
Through analysis of exosomal cfDNA in urine or blood, biomarkers for the early diagnosis of liver diseases, cardiovascular diseases, and neurodegenerative diseases can be discovered.
Organ Transplant Monitoring
This service can be used to identify exosomal cfDNA associated with organ damage or rejection in transplanted organs.
Gut Microbiome Research
Exosomal cfDNA can provide genomic information about the gut microbiota, aiding in research on the relationship between dysbiosis and metabolic diseases, intestinal inflammation, and other conditions.
FAQ
Q. How to choose the appropriate cfDNA analysis method (NGS vs. qPCR vs. ddPCR)?
The choice of cfDNA analysis method depends on the research objectives and sample characteristics:
1. NGS (High-Throughput Sequencing): Suitable for whole-genome or targeted mutation screening, NGS can detect point mutations, copy number variations (CNVs), structural variations, and methylation modifications. However, it requires a higher DNA input and is more expensive.
2. qPCR (Real-Time Quantitative PCR): Suitable for the quantitative detection of known mutations, qPCR offers lower sensitivity but is cost-effective and easy to operate.
3. ddPCR (Digital PCR): Ideal for detecting low-abundance mutations (such as tumor micro-clones) due to its extremely high sensitivity, ddPCR can detect allele mutations below 0.1%. It is especially well-suited for tumor biomarker and trace cfDNA analysis.
Selection Strategy: For comprehensive mutation analysis, choose NGS. For high-sensitivity mutation detection, use ddPCR. For quantifying specific, known mutations, qPCR offers a cost-effective option.
Q. How to optimize the extraction of exosomal cfDNA to ensure high-quality analysis?
1. Select an appropriate lysis buffer:
Ensure complete release of exosomal cfDNA while preventing degradation.
2. Improve cfDNA recovery:
Use magnetic beads or silica column-based methods to enhance cfDNA recovery and remove protein and RNA contaminants.
3. Fragment analysis:
Utilize a Bioanalyzer or Qubit to assess cfDNA fragment sizes, ensuring that the main fragments are within the 100-500 bp range to avoid contamination from high-molecular-weight genomic DNA.
4. Prevent degradation:
Employ RNA/DNase inhibitors throughout the process, store samples at -80°C, and avoid repeated freeze-thaw cycles.
Case Study
This study used digital PCR (ddPCR) to accurately quantify the KRAS mutation status in exosomal DNA. The results showed that exosomal DNA carrying KRAS mutations is closely associated with poor prognosis in metastatic colorectal cancer patients, with its presence indicating shorter overall survival and progression-free survival. These findings suggest that exosomal KRAS mutation status may serve as an important biomarker for predicting treatment efficacy and patient prognosis.
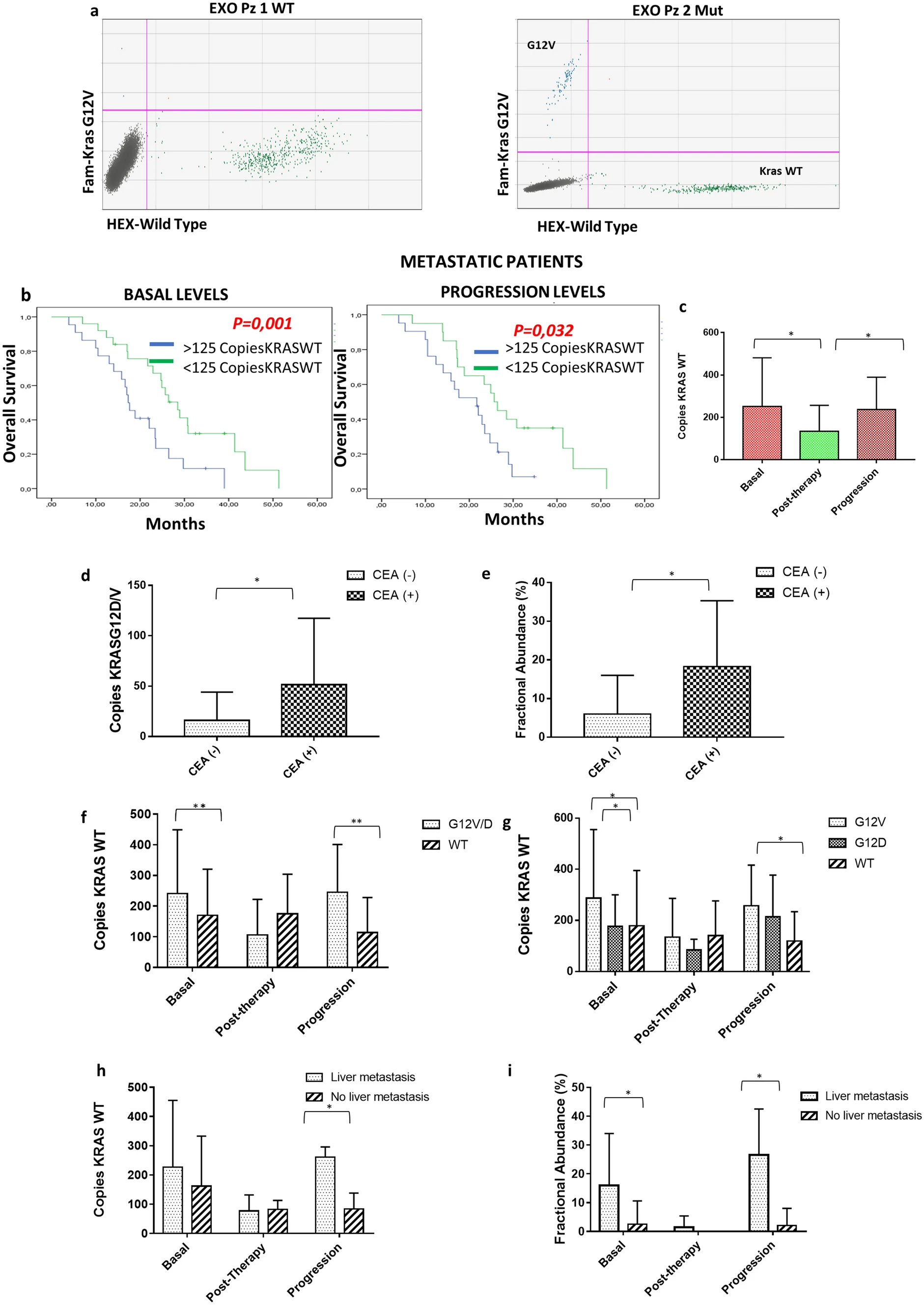
Lucchetti D. et al. Nature Publishing Group. 2021.
How to order?







