Edman Degradation Protein Sequencing service
Edman degradation protein sequencing is a sequencing technology that uses chemical methods to cut amino acids one by one from the N-terminus of proteins or peptide chains and identify them. Compared with mass spectrometry, Edman degradation has unique advantages in determining the N-terminal sequence of proteins. It can directly and step by step analyze the amino acid sequence from the N-terminus of proteins, ensuring the high accuracy and reliability of sequencing results. Edman degradation protein sequencing service is suitable for N-terminal sequence determination of purified proteins and peptides, and can provide key sequence information for subsequent functional studies and structural analysis. MtoZ Biolabs provides Edman degradation protein sequencing service which can determine the N-terminal sequence of purified protein products, antibodies, protein vaccines, etc.
Technical Principles
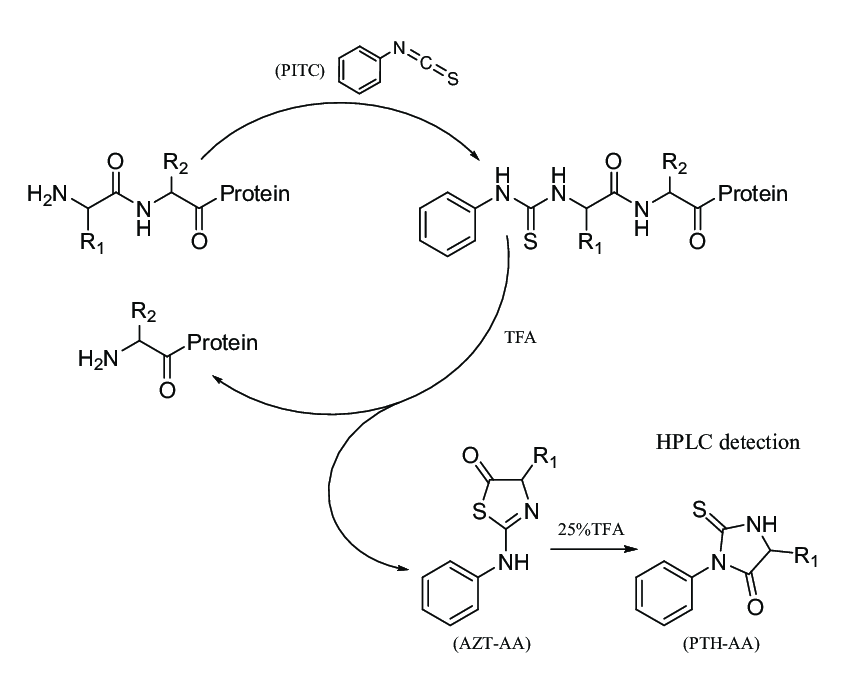
Yang W. et al. Molecules. 2021.
Edman degradation protein sequencing service employs a chemical reaction to sequentially cleave and identify amino acid residues from the N-terminal of a protein or peptide chain. Phenylisothiocyanate (PITC) reacts with the N-terminal amino acid to form a phenylthiocarbamoyl (PTC)-amino acid. Under acidic conditions, the N-terminal PTC-amino acid is selectively cleaved to form a stable phenylthiohydantoin (PTH)-amino acid. The released PTH-amino acids are then separated and identified using high-performance liquid chromatography (HPLC), allowing the N-terminal amino acid sequence of the protein or peptide chain to be deduced step by step.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Edman degradation protein sequencing service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Edman degradation protein sequencing service data, providing clients with a comprehensive data report.
4. Complete N-terminal sequencing service: Using a specific protein loading system, MtoZ Biolabs can measure the 60-70 amino acids at the N-terminus. In addition to the Edman degradation protein sequencing service, we also provide mass spectrometry protein sequencing service, which can measure blocked and modified protein ends, complementing the Edman degradation method.
Sample Submission Suggestions
Sample Types
Purified proteins, peptides, and protein samples transferred onto PVDF membranes.
Sample Quantity
We recommend providing at least 10-50 pmol of protein samples with >90% purity.
Specific requirements may vary depending on project requirements. Please contact us for consultation before sample submission.
FAQ
Q. How to Overcome the Limitation of Cycle Numbers in Edman Degradation protein sequencing service?
1. Optimization of Degradation Conditions
Precisely control parameters such as pH, temperature, and reaction time to enhance the efficiency of each Edman degradation cycle. Use HPLC to optimize separation conditions and minimize interference from undegraded byproducts.
2. Sample Preparation
Ensure high purity of the sample during pre-treatment to reduce background noise that may interfere with the degradation process. Select stable peptides or shorter fragments for degradation.
3. Integration with Enzymatic Digestion
Use specific proteases such as trypsin, Lys-C, or Glu-C to cleave proteins into shorter peptides. Perform Edman degradation on each peptide fragment individually. Use bioinformatics tools to assemble the peptide sequences into a complete N-terminal sequence.
4. Combined with Mass Spectrometry
For sequences that cannot be fully resolved by Edman degradation, supplement the data with LC-MS/MS analysis. Mass spectrometry provides broader sequence coverage and can compensate for Edman degradation's cycle limitations.
While Edman degradation has an inherent cycle number limitation, optimizing degradation conditions, employing enzymatic digestion, and integrating mass spectrometry can effectively extend sequence coverage, enabling the determination of longer and more complete protein N-terminal sequences.
Case Study
Edman degradation protein sequencing was employed to analyze the n-terminal amino acid sequence of the vaccine core antigen, confirming a high degree of consistency between the n-terminal structure of the vaccine protein and its intended design. This provides a solid foundation for protein quality assurance, supporting subsequent animal studies and Phase I clinical trials for immunological evaluation.
Yang J. et al. Nature Communications. 2024.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







